Synthesis Mechanism of 21st Amino Acid Selenocysteine (Press Release)
- Release Date
- 05 Apr, 2013
- BL41XU (Structural Biology I)
RIKEN
The University of Tokyo
Key points
• The three-dimensional structure of SelA, the enzyme required for selenocysteine (Sec) synthesis, is determined.
• Four subunits of the huge star-shaped protein, SelA, play different roles in Sec synthesis on one tRNA.
• The findings will contribute to the development of superenzymes by facilitating introduction of selenium (Se) into enzymes.
|
A group from RIKEN (president, Ryoji Noyori) and the University of Tokyo (president, Junichi Hamada) clarified the synthesis mechanism of Sec, which is the “21st amino acid”*1 in bacteria. This was achieved by a joint research group of Shigeyuki Yokoyama (director, currently senior scientist of Structural Biology Laboratory) of the Systems and Structural Biology Center at RIKEN, Yuzuru Itoh (assistant professor) of the Institute of Molecular and Cellular Biosciences, the University of Tokyo, and scientists from Yale University, USA. Se is an element effective for preventing aging and lifestyle-related diseases and is essential as a micronutrient in various living organisms including humans. Se deficiency causes cancers and hypertension. In vivo, Se exists in Sec and functions when incorporated into some proteins in accordance with the genetic code that determines amino acid sequences. Sec is synthesized from another amino acid, which is bound to Sec-specific transfer RNA (tRNA)*2, tRNASec; however, the synthesis mechanism of Sec differs between (1) the group of eukaryotes*3, including humans, and archaea*4 (human type) and (2) the group of bacteria*5 (bacteria type). While the synthesis mechanism in the human type has been clarified, that of bacteria has not been elucidated. The joint research group analyzed the crystal structure of the complex of SelA, the bacterial Sec synthase, and tRNASec, and found that SelA forms a large star shape consisting of ten subunits.*6 They also examined the detailed functions of each subunit and found that the star-shaped structure is essential for Sec synthesis. These findings will significantly contribute to the development of methods for synthesizing proteins containing Se. This will enable the development of superenzymes with functions superior to those of natural enzymes, as well as promote research on diseases caused by Se deficiency. This research was supported by the Targeted Proteins Research Program and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology. The findings were published in the American scientific journal Science on 5 April 2013. Publication: |
<<Figures>>
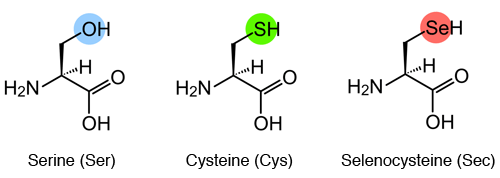
Sec is an amino acid in which the oxygen (O) of Ser or the sulfur (S) of Cys is replaced by Se.
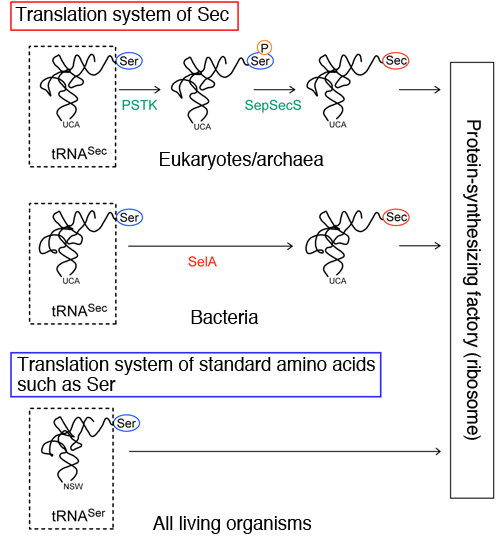
Top: Translation system of Sec in eukaryotes and archaea. Ser is converted to Sec by two enzymes, PSTK and SepSecS, in two steps. First, PSTK discriminates Ser-tRNASec and marks the Ser by transferring a phosphate group (P) to the Ser. Then, SepSecS converts only the marked Ser to Sec.
Middle: Translation system of Sec in bacteria. Ser is converted to Sec by one enzyme, SelA, in one step.
Bottom: Translation system of standard amino acids.
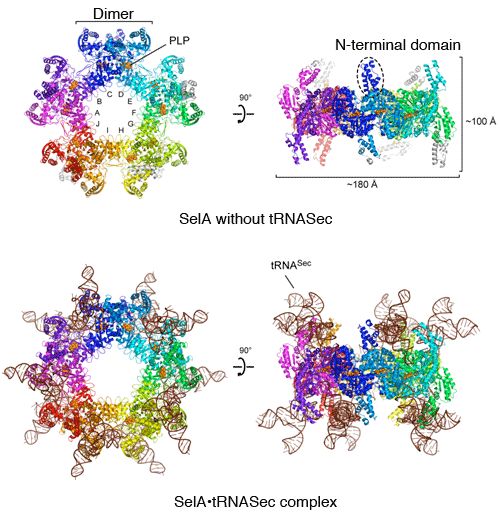
Top: SelA without tRNASec. Front (left) and side (right) views of star-shaped SelA. The N-terminal domain protrudes from the star-shaped platelike structure. The ten subunits form a ring and the ten catalytic sites binds the coenzyme pyridoxal 5′-phosphate (PLP), which helps SelA activity. PLP is a derivative of vitamin B6, which is transformed in the body and helps the activity of various enzymes.
Bottom: SelA·tRNASec complex. Front (left) and side (right) views. Ten tRNAsSec bind to SelA.
(YouTube: Animation of overall structure of SelA·tRNASec complex)
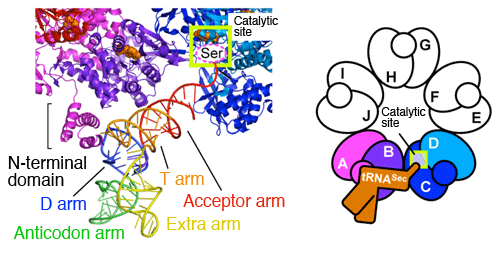
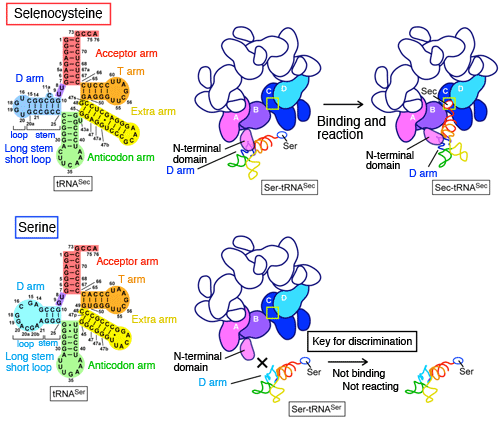
Enlarged view of tRNASec binding to subunits A and B of SelA. The N-terminal domain of subunit A interact with the unique D arm of tRNASec and discriminates tRNASec from other tRNAs. Four subunits (A to D) play different roles in cooperation with each other for one Ser-tRNASec; (1) subunit A discriminates Ser-tRNASec; (2) subunits A and B fix Ser-tRNASec; (3) subunit C catches the end of Ser-tRNASec; and (4) subunits C and D convert the Ser at the end of Ser-tRNASec to Sec in one step. Because all subunits have the same structure, subunit C plays the role of (1), subunits C and D play that of (2), subunit E plays that of (3), and subunits E and F play that of (4) for the next Ser-tRNASec. For the Ser-tRNASec on the opposite side, subunit D plays the role of (1), subunits D and C play that of (2), subunit B plays that of (3), and subunits B and A play that of (4). Each subunit can play any of the four roles.
Top: Mechanism of tRNASec discrimination. The N-terminal domain of SelA interacts with the D-arm structure, which is unique to tRNASec and converts the Ser to Sec on the tRNASec. The D arm of tRNA consists of a stem and a loop, and the D arm of tRNASec is characterized by a long stem and a short loop.
Bottom: Mechanism of tRNASer discrimination. The D arm of tRNA for standard amino acids such as tRNASer consists of a short stem and a long loop. Because of this structure, the D arm of tRNASer cannot bind to SelA and therefore the Ser attached to tRNASer is not converted to Sec.
<<Glossary>>
*1 21st amino acid
Generally, proteins consist of 20 kinds of amino acid that are joined together to form a chainlike structure. Amino acid sequences are determined by the genetic code of DNA base sequences. The conversion of the base sequences to amino acid sequences is called “translation”. On the ribosome, amino acids are joined together in the order specified by messenger RNA (mRNA), which is the transcript of DNA. There are four bases that make up mRNA, namely, A, U, G, and C, and a sequence of three of these bases is called a codon. There are 64 (4×4×4) codons, of which 61 encode any of the 20 amino acids. For example, UCA and AGU codons encode serine (Ser). The remaining three codons are stop codons that terminate the amino acid sequences. Sec is an amino acid existing in some animals including humans, plants, protozoa, archaea and bacteria. Because Sec is a newly found amino acid in addition to the standard 20 amino acids, it is called the “21st amino acid”. The codon for Sec (UGA codon) is a stop codon but encodes Sec only when there is a specific sequence on mRNA.
*2 Transfer RNA (tRNA)
tRNA is a type of RNA in which 70 to 100 bases (A, U, G, and C) are joined together in a chainlike structure. There is one or more tRNAs for each amino acid. tRNA is attached with a specific amino acid through its end and carries the amino acid to a ribosome. tRNA consists of five regions, namely, the acceptor arm, D arm, anticodon arm, extra arm, and T arm. Each arm has a region called a stem forming a double helical structure, in which the facing bases are paired with each other, and a region called a loop not forming a double helical structure. An amino acid binds to the end of the acceptor arm, and the anticodon, the three bases at the end of the anticodon arm, forms base pairs with a codon on mRNA in a ribosome. The D and T arms three-dimensionally interact with each other to form an L-shaped structure, from which the extra arm protrudes.
*3 Eukaryotes
Eukaryotes are a group of living organisms that have an organelle called a nucleus containing DNA in their cells. Animals including humans, fungi, plants and protozoa belong to this group.
*4 Archaea
Archaea are a group of microorganisms that are also called the archaebacteria. Archaea are one of the three major groups of living organisms along with eukaryotes and bacteria. Archaea are different from eukaryotes in that they have no cellular nucleus, but are significantly similar to eukaryotes in terms of basic systems such as the synthesis mechanism of amino acids and proteins. Archaea are more closely related to eukaryotes than to bacteria. As observed for methanogens, halophiles and hyperthermophiles, archaea often live in extreme environments.
*5 Bacteria
Bacteria are a group of microorganisms that are also called eubacteria. They have no cellular nucleus. Escherichia coli, Bacillus subtilis, Lactobacillus, and Salmonella are examples of bacteria.
*6 Subunits
Proteins are categorized into monomeric and multimeric ones. Monomeric proteins are consisting of a single polymer chain of amino acids (polypeptide chain), while multimeric ones consisting of multiple polymer chains. Each polypeptide chain that constitutes a multimeric protein is called a subunit.
|
For more information, please contact: |
- Current article
- Synthesis Mechanism of 21st Amino Acid Selenocysteine (Press Release)