Structure of the human DNA recombinase reveals a double ring- Homologous pairing takes place at the central channel of the ring - (Press Release)
- Release Date
- 07 May, 2004
- BL44B2 (RIKEN Materials Science)
7 May 2004
RIKEN
|
RIKEN (President Ryoji Noyori) has determined the atomic resolution structure of the human Dmc1 protein, and succeeded in providing the first clear picture of how Dmc1 promotes homologous recombination. This work was done by the members of the Protein Research Group (Dr. Shigeyuki Yokoyama, Project Director, Dr. Takashi Kinebuchi, Research Scientist, Dr. Hitoshi Kurumizaka, Visiting Research Scientist) at the Genomic Sciences Center (Dr. Yoshiyuki Sakaki, Director). |
1. Background
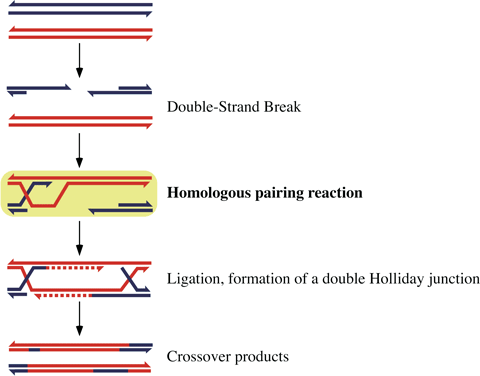
Meiosis is a cell division process specifically occurring in germ cells (testis and ovary) of eukaryotes. During the cell division, homologous chromosomes pair and parts or all of genes are shuffled between the chromosomes, a process called homologous recombination (Figure 2). Eukaryotes obtain genetic variation by this method. Homologous recombination is also essential for the repair of DNA damages; thus for the cell to function properly, homologous recombination is important.
The members have focused on the protein that is responsible for the homologous recombination in germ cells, namely Dmc1, and has succeeded in crystallizing the full-length human protein. The crystal was used to determine the three-dimensional structure of Dmc1 at atomic resolution. Based on the structure, the research group has studied the homologous recombination promoted by Dmc1.
2. Methods
The human Dmc1 protein, consisting of 341 amino acids, binds both single-stranded DNA and double-stranded DNA, and promotes the homologous-pairing reaction, a key step of homologous recombination. The members overexpressed the Dmc1 protein in E. coli and purified the Dmc1 protein using several chromatographic techniques. A single crystal (100 mm x 600 mm x 600 mm) of Dmc1 was obtained (Figure 3), and the crystal structure of Dmc1 was successfully determined by using the data collected at the synchrotron radiation of the RIKEN Structural Biology II Beamline (BL44B2) at SPring-8.
3. Results
1) Dmc1 forms a double-ring structure composed of 16 monomers
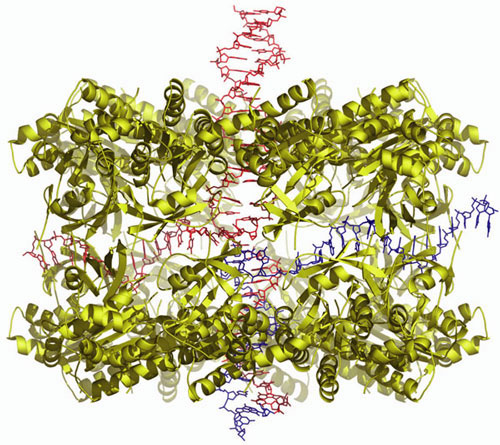
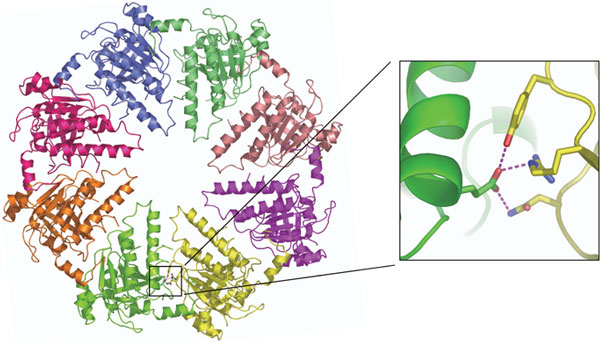
The human Dmc1 protein is a homolog of the E. coli RecA protein. Studies have shown that the bacterial RecA protein forms a helical filament structure and promotes homologous pairing. In contrary to the extensive sequence similarity between Dmc1 and RecA, the members show that the Dmc1 functions as a double-ring structure. The ring consists of 8 subunits and two rings are stacked in a bi-polar nature with a diameter of 130 Å and a height of 85 Å (Figure 5).
2) Why does Dmc1 form a stable ring structure?
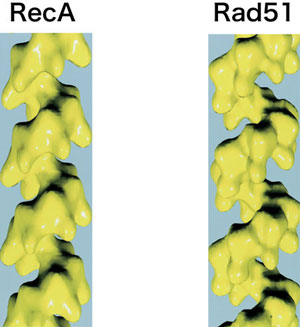
Dmc1 has a homolog called Rad51, which exists in both mitotic and meiotic cells (Figure 4). Interestingly, Rad51 forms a helical filament structure that closely resembles the RecA filament structure, despite the close amino acid sequence similarity between Dmc1 and Rad51. What determines this structural difference? The present study identifies an amino acid residue of Dmc1 that makes three hydrogen bonds at the subunit-subunit interface. In Rad51, a different amino acid residue is present at the corresponding location that is unlikely to form hydrogen bonds. The members show that these hydrogen bonds in Dmc1 are essential for stabilizing the octameric ring structure (Figure 6).
3) A model of the recombination intermediate complex
The Dmc1 double-ring structure contains two open-ended passages: one at the center of the ring (about 30 Å in diameter) and the other between the stacked rings (about 15 Å in diameter). Based on alanine-scanning mutagenesis studies, the central channel was shown to bind double-stranded DNA, while the passage at the side of the ring that leads to the central channel was shown to bind single-stranded DNA. These results led to the model of the ternary complex containing Dmc1, single-stranded DNA, and double-stranded DNA. In this model, homologous pairing takes place at the center of the double ring, and the recombined DNA is spooled out of the passages. This mechanism contrasts with those proposed for the helical filament forming RecA and Rad51 proteins. Hence, the present studies suggest that there are at least two recombination mechanisms utilized in germ cells. Such multiple mechanisms may be essential for cells to properly divide during meiosis.
4. Perspectives
Since homologous recombination is the underlying mechanism of gene therapy and genetic engineering, understanding the structural and biochemical properties of eukaryotic recombinases is essential for improving these technologies. The present study may provide a foundation for modifying the activity of the Dmc1 recombinase, leading to an increased recombination frequency in eukaryotic cells. Therefore, the structural information of Dmc1 recombinase may contribute to the advancement of gene therapeutic and transgenic technologies.
<<Figures:>>
The search and the pairing of two homologous sequences take place at the center of the Dmc1 double ring. The dsDNA is shown entering from one end of the stacked ring, and the paired product is shown coming out from the other end of the stacked ring. The ssDNA is shown passing through the cavity created by the stacking of two Dmc1 rings.
In contrast to the ring structure of Dmc1 , these two proteins form helical filament structures
The Dmc1 double-ring structure viewed from the top (left) and from the side (right).
The closeup view of the tripartite hydrogen bonds located at the subunit-subunit interface is shown on the right box. These hydrogen bonds (colored in pink) are present in each subunit-subunit interface.
|
For further information, please contact: Takashi Kinebuchi for SPring-8, |
- Previous Article
- Successful Biaxial Time-Sharing Mapping of Intramolecular Motion in Single Protein Molecules (Press Release)
- Current article
- Development of a methodology to control polymer orientation - improving solar cell efficiency (Press Release)