Clarifying Rhythmic Structural Changes in Cyanobacterial Clock Protein (Press Release)
- Release Date
- 27 Nov, 2010
- BL45XU (RIKEN Structural Biology I)
Japan Science and Technology Agency (JST)
Nagoya University
RIKEN
|
As part of JST Use-Inspired Fundamental Research, Takao Kondo (professor) and Shuji Akiyama (lecturer) of the Graduate School of Science, Nagoya University, and their coworkers have clarified that a clock protein*1of cyanobacteria has a 24 h cycle involving rhythmic expansion and contraction similar to a heart beating. Cyanobacteria are the simplest known form of life with a circadian clock, which comprises three types of clock protein (KaiA, KaiB, and KaiC). Scientists of this research group previously demonstrated that when these clock proteins were mixed in the presence of ATP,*2 the ATPase activity of KaiC and the phosphorylation state change with a period of 24 h. This indicates that the ATPase activity of KaiC is a significant factor in determining the period of circadian clocks. It was hoped to clarify how the molecular structure of KaiC changes with increasing or decreasing ATPase activity. Scientists in this research group clarified that the shape of a KaiC molecule in solution rhythmically changes with a period of 24 h by means of fluorescence spectroscopy and using RIKEN Structural Biology I Beamline (BL45XU, small-angle X-ray scattering*3) at SPring-8. KaiC has a double-ring structure similar to a stack of two donuts: the radius of one ring repeatedly increases and decreases while closely associated with the controlled state of ATPase in the other ring. KaiA and KaiB sense these expansions and contractions and use them for the timing of their assembly with and disassembly from KaiC. KaiC converts the reference signals of ATPase into structural changes and couples the changes with the assembly with and disassembly from KaiA and KaiB, realizing dynamic and stable circadian ticking. In this research, structural changes of KaiC were partly clarified, and a molecular technological basis clarifying the mechanism underlying the circadian ticking of KaiC through the self-control of its ATPase activity was established. It is hoped that the framework of research on molecular clocks using ATP, which is being clarified through the investigation of cyanobacteria, will provide clues leading to the clarification of circadian clocks in higher organisms including humans. The achievements of this research were published as an advanced online publication in the European scientific journal The EMBO Journal on 26 November 2010. (Publication) |
《Glossary》
*1 Clock protein
Clock protein is the collective term for proteins essential for maintaining the circadian clock of living organisms. Mutations and deficiencies in clock proteins modulate the rhythms of various activities of living organisms.
*2 ATP
Adenosine triphosphate (ATP) is used as an energy source in various reactions including muscle contraction. It is a nucleotide used for the conservation and consumption of energy in living organisms. It is also called the “energy currency” because of its presence in significant amounts and its importance in the metabolism of living organisms. Three phosphate groups bind to an adenosine. In the presence of ATPase, phosphate groups are separated and ATP is degraded. Approximately 8 kcal/mol of energy is released when a phosphate group is separated.
*3 Small-angle X-ray scattering
Small-angle X-ray scattering is used to obtain structural information on a substance by irradiating X-rays onto a sample and detecting the X-rays that scatter at small angles. It is useful for the nanometer-scale structural analysis of, for example, internal structures of biological polymers such as proteins and nucleic acids, microparticles, liquid crystals, and alloys.
《Figure》
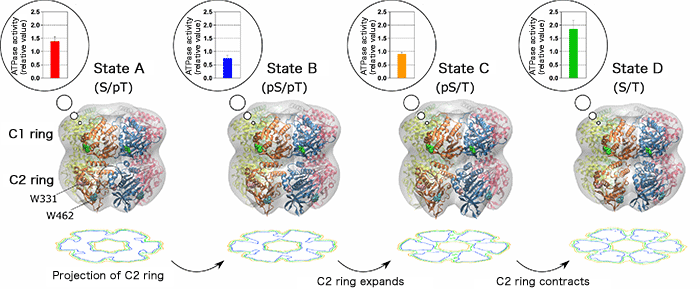
The gray region represents the shape of the KaiC molecule determined by small-angle X-ray scattering. The ribbon model is a high-resolution molecular model constructed by optimizing the known X-ray crystal structures so as to coincide with the small-angle X-ray scattering data. W331 and W462 represent the tryptophan residues in the C2 ring. Contour plots obtained by projecting the C2 ring alone are shown below each model. The radius of the C2 ring changes negligibly after the transition from state A to state B. After the transition from state B to State C, however, the radius of the C2 ring greatly increases, as indicated by the deepened trough of the counter plot. The expansion and contraction of the C2 ring are coupled with the increase and decrease in the ATPase activity of the C1 ring, respectively.
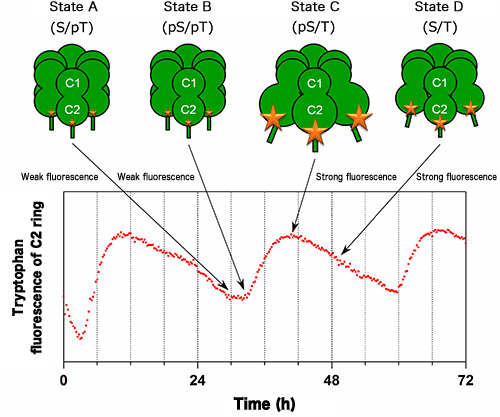
Changes in the intensity of fluorescence from tryptophan residues (W331 and W462 in Fig.1) in the C2 ring of KaiC were measured in the presence of KaiA and KaiB. The size of the star marks on KaiC corresponds to the intensity of fluorescence from tryptophan residues. The intensities of fluorescence from W331 and W462 also increase and decrease with a period of 24 h in accordance with the expansion and contraction of the C2 ring shown in Fig.1, respectively.
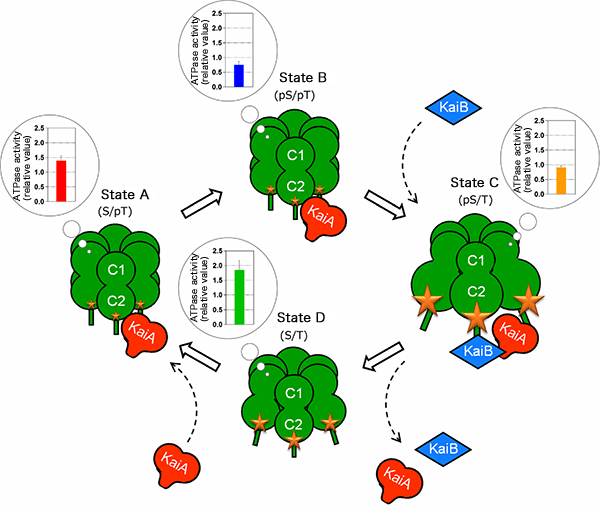
The controlled state of the ATPase in the C1 ring is associated with the expansion or contraction state of the C2 ring. KaiA and KaiB use this “molecular beating” as a guide for the timing of their assembly with and disassembly from KaiC. KaiA and KaiB bound to KaiC modulate the ATPase activity, stabilizing the reaction cycle.
|
For more information, please contact: |
- Previous Article
- First-Ever Success in Crystal Structure Clarification of Enzyme That Catalyzes Nitrous Oxide, a Greenhouse Gas, Generation (Press Release)
- Current article
- Clarifying Rhythmic Structural Changes in Cyanobacterial Clock Protein (Press Release)