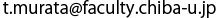Elucidation of the Rotation Mechanism of the Molecular Motor, Which is Implicated in Osteoporosis and Cancer Metastasis (Press Release)
- Release Date
- 14 Jan, 2013
- BL41XU (Structural Biology I)
Chiba University
Japan Science and Technology Agency
RIKEN
Kyoto University
|
Associate professor Takeshi Murata (PRESTO researcher at JST, visiting researcher at RIKEN) and others revealed the detailed structure of the V1-ATPase molecular motor (a protein nanomotor NOTE1) for the first time in the world using the high-intensity X-ray available at SPring-8. The research results made clear, on an atomic level, the general framework of the energy conversion process, from ATP energy to rotational motion. The information obtained through this research is expected to play an important role in predicting the method to inhibit V-type ATPase activity, implicated in such diseases as osteoporosis and cancer, leading to drug discovery based on the knowledge of steric structure. This research was conducted under the auspices of several funding schemes including Targeted Protein Research Program (MEXT), Grant-in-Aid for Scientific Research (MEXT) and JST Strategic Basic Research Programs (PRESTO), and in collaboration with Prof. Ichiro Yamato (Graduate School of Industrial Science and Technology, Tokyo University of Science), Prof. So Iwata (Graduate School of Medicine, Kyoto University), Prof. Shigeyuki Yokoyama (RIKEN Systems and Structural Biology Center) and others. The research results were published in the online version of Nature (a UK science journal) on the 13th of January, 2013 (at 18:00 UKT). Publication: |
<<Figures>>
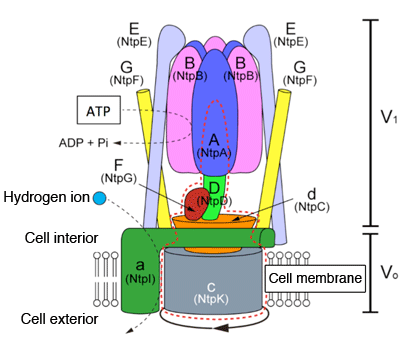
The V-type ATPase is a supermolecular complex generally consisting of nine to thirteen proteins, and its structure is divided into two parts: water soluble protein portion (V1) and membrane protein portion (Vo). The names shown in parentheses represent the old nomenclature used for the V-ATPase subunit of Enterococcus hirae. ATP is hydrolyzed by the catalytically-active head (A3B3), and the resultant energy is used to rotate the rotary shaft (DFd) and rotor ring (c), and for transporting hydrogen ions out of the cell (or into the organelle).
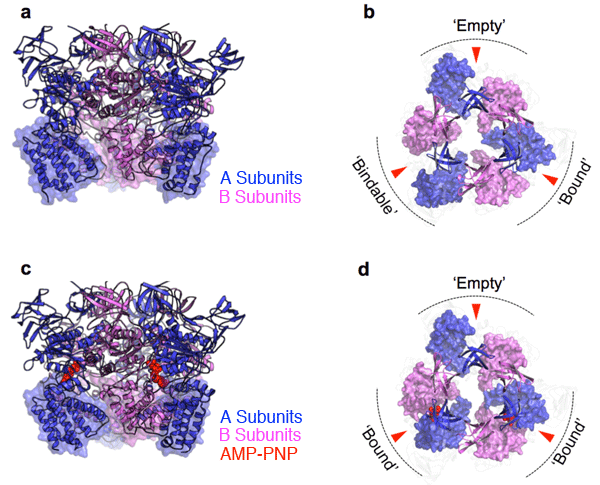
a) A3B3 complex crystal structure (side view)
b) A3B3 complex crystal structure (top view). For easier viewing, only the N-terminal β barrel domains and C-terminal domains are shown. ATP binding sites (three in all) are indicated by red arrows.
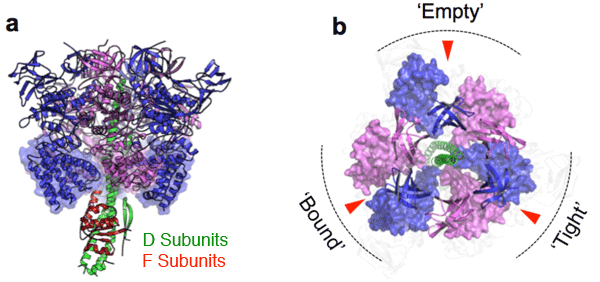
c), d) AMP-PNP bound A3B3 complex crystal structure
Shown in the same format as in a) and b).
a) V1-ATPase crystal structure (side view)
b) V1-ATPase crystal structure (top view). For easier viewing, only the N-terminal β barrel domains, C-terminal domains, and D subunits are shown. ATP binding sites (three in all) are indicated by red arrows.
<<Glossary>>
NOTE1 Protein nanomotor
The generic nomenclature for the protein complexes - typically of a size measured by nanometers (10-9m) - that convert biological energy obtained through hydrolysis of ATP within biological cells into mechanical motion.
|
For more information, please contact: |
- Previous Article
- Discovery of Nano-Porous Materials Capable of Memorizing/Erasing Molecule Adsorption Information (Press Release)
- Current article
- Elucidation of the Rotation Mechanism of the Molecular Motor, Which is Implicated in Osteoporosis and Cancer Metastasis (Press Release)