Suppression of Amplification Errors during Polymerase Chain Reaction (PCR) (Press Release)
- Release Date
- 09 Apr, 2013
RIKEN
Key points
• A technique to suppress nonspecific amplification and DNA mutations, both of which are inevitable in DNA amplification, was developed.
• A simple technique that only requires addition of a protein to the reaction solution was developed.
• The findings are expected to contribute to the improvement of the accuracy of DNA handling techniques applied in various fields.
|
A group from RIKEN (president, Ryoji Noyori) succeeded in improving the accuracy of PCR*1, which is indispensable to genetic diagnosis and genomic analysis, using a protein involved in the DNA mismatch repair system*2 inherent to organisms. This achievement was realized through the Structural-Biological Whole-Cell Project of Thermus thermophilus HB8*3 by a research group led by Kenji Fukui (visiting scientist; currently, Office for University-Industry Collaboration, Osaka University), Yoshitaka Bessho (team leader), and Seiki Kuramitsu (group director) of the Synchrotron Radiation System Biology Research Group, RIKEN SPring-8 Center (director, Tetsuya Ishikawa). DNA is called the genetic blueprint of organisms and has been used in various applications in modern society from basic biological research to clinical diagnosis and criminal investigation using the DNAs in residual materials. In the analysis of DNAs, PCR is indispensable for amplifying the amount of the desired DNA sequence to a predetermined amount. In PCR, only the desired region from a long DNA sequence is amplified by approximately a billionfold using an extremely small amount of template DNA as a starting material to facilitate detection and analysis of the desired DNA. During PCR, DNAs are amplified in every heating (approximately at 95°C) and cooling (approximately at 50°C) cycle. However, there are two types of PCR-related error, (1) amplification of undesired DNA and (2) the amplification of desired but mutated DNA. The research group has analyzed the structures and functions of various proteins that function even in environments with temperatures exceeding 90°C in the Structural-Biological Whole-Cell Project of Thermus thermophilus HB8; in particular, DNA mismatch repair proteins were found to recognize DNA mismatches extremely specifically and bind to the mismatched pairs. The research group focused on the finding that mismatched base pairs*4, a type of DNA error, are formed during the occurrence of the above-mentioned two PCR-related errors, and considered applying MutS, a DNA mismatch repair protein that strongly binds to the mismatched base pairs. When MutS was added during PCR to amplify a specific DNA region using DNA of a bacterium, nonspecific amplification was suppressed depending on the amount of MutS added, and the number of mutations was reduced to approximately one-third. These achievements are expected to improve the efficiency and accuracy of PCR-related technologies applied in various fields. The results of this study were published in a Swiss scientific journal, the International Journal of Molecular Sciences, on 22 March 2013. Publication: |
<<Figures>>
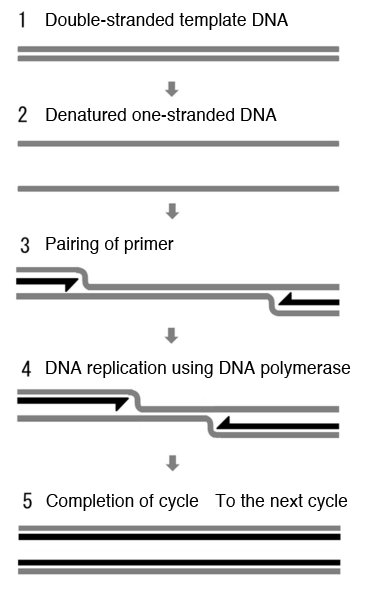
1 and 2. Double-stranded template DNA is heated and denatured to single-stranded DNAs.
3. Short single-stranded DNA called the primer (indicated by black arrows) preferentially pairs with the template DNA upon cooling.
4. DNA polymerase recognizes the hybrid of the primer and template DNA and starts DNA replication.
5. The obtained double-stranded DNAs are used as a template DNA in the next cycle.
DNAs are further amplified by repeating the above cycle.
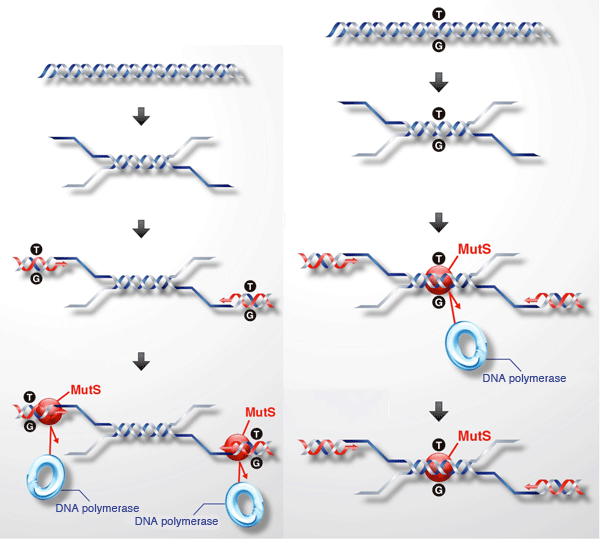
MutS (red circle) strongly binds to a mismatched base pair (green and yellow symbols).
Left: When a primer used to specify the region of amplification erroneously binds to a nonspecific region, a mismatched base pair (indicated by T/G) forms. When the mismatched base pair is bound by MutS (red circle), the DNA polymerase used to amplify DNAs cannot bind to the mismatched base pair, thus suppressing the amplification of the undesired DNA sequence.
Right: Mismatched base pairing also occurs when the DNA polymerase amplifies a desired but mutated sequence. MutS strongly recognizes such a region, suppressing the function of DNA polymerase to inhibit the further amplification of DNA containing a mutated sequence.
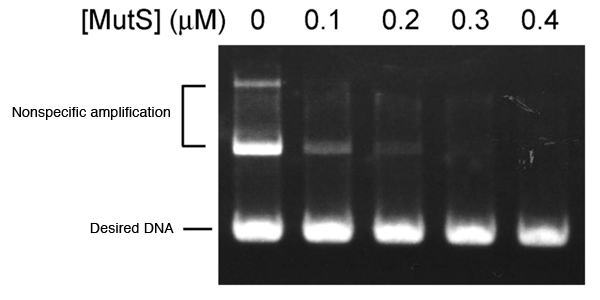
DNA product is segmented according to its length and visualized after PCR. In the absence of MutS (left), two types of nonspecific amplification are observed above the desired DNA. Nonspecific amplification is not observed with increasing amount of MutS added.
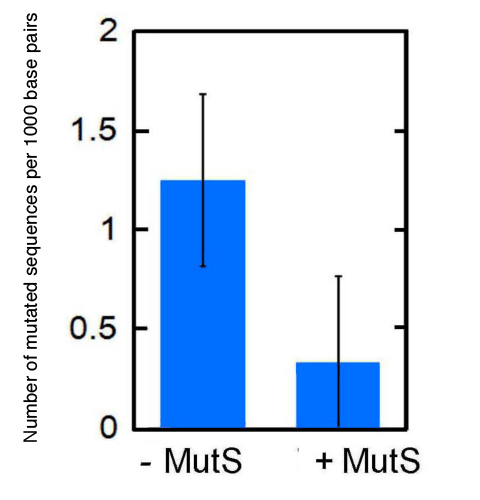
The DNA sequence of the fragments amplified by PCR is confirmed and the number of mutated sequences per 1000 base pairs is plotted on the ordinate. With addition of MutS, the number of mutated sequences is reduced to approximately one-third. The experiments were performed five times. Bars indicate standard deviations.
<<Glossary>>
*1 PCR
PCR is a technique used to amplify DNA sequences. Within a DNA template, only the desired region is amplified by PCR. Even a small amount of a template can be amplified by approximately a billionfold by PCR. The PCR is as follows. The double-stranded DNA used as a template is denatured at a high temperature to generate single-stranded DNAs. When an excessive amount of short DNA molecules called primers is added to the reaction solution while it is cooled, the primers preferentially bind to the template to form double-stranded DNAs. DNA polymerase, which catalyzes DNA synthesis, recognizes the double-stranded DNA region consisting of the template and primer, and synthesizes complementary DNAs for the remaining single-stranded region starting from the end of the primer. Thus, double-stranded DNAs are replicated. DNAs are further amplified by repeating the above cycle using the replicated DNAs as the templates. DNA polymerase should be added in each cycle if DNA polymerase is also denatured at high temperatures required to denature DNA. However, reaction is continued without adding DNA polymerase when extremely thermophilic organism-derived heat-resistant DNA polymerase is used.
*2 DNA mismatch repair system
DNAs are constantly damaged by internal factors, such as replication errors and the presence of active oxygen species, as well as external factors, such as radiation of ultraviolet rays and ionizing radiation. However, organisms have various DNA repair mechanisms to overcome such damage and various DNA mismatch repair proteins are involved in these mechanisms.
*3 Structural-Biological Whole-Cell Project of Thermus thermophilus HB8
This research project is conducted mainly by the researchers of Osaka University in cooperation with many researchers of other institutions. The project aims to establish an academic foundation that leads to the systematic understanding of every life phenomenon at the cellular level on the basis of the structures and functions of DNA, proteins, carbohydrates, lipids, and other low-molecular-weight compounds, using Thermus thermophilus HB8 as the model representing all the organisms on the earth. The project is divided into the following four stages.
First stage: Analysis of the 3D structure of a whole cell consisting of molecules such as a protein
Second stage: Functional analysis of a whole cell consisting of molecules such as a protein
Third stage: Analysis of various systems (network relationship among several molecules) in a cell
Fourth stage: Simulation of the whole cell
*4 Mismatched base pairing
Each of four bases, i.e., adenine (A), guanine (G), cytosine (C), and thymine (T), on one strand bond with one type of base on the other strand (A-T and C-G) to form a double-stranded DNA. Other types of pair, e.g., G-T and C-A, are called mismatched base pairs. Mismatched base pairing is caused when the DNA polymerase takes in mismatched bases during DNA replication by the proliferation of cells in organisms. Almost all organisms including humans have a mismatch repair system, which recognizes and repairs mismatched base pairs, improving the accuracy of DNA replication.
|
For more information, please contact: |
- Previous Article
- Elucidation of the molecular vibration of the high-valent non-heme diiron enzyme intermediates (Press Release)
- Current article
- Suppression of Amplification Errors during Polymerase Chain Reaction (PCR) (Press Release)