World's first determination of steric structure of transient electron-transfer complex formed when copper-containing nitrite reductase reacts with electron-donor protein during denitrification (Press Release)
- Release Date
- 05 Nov, 2009
- BL44XU (Macromolecular Assemblies)
This result is expected to be helpful for the development of strategies to reduce the global amount of harmful nitrogen oxides
Osaka University
|
A research group consisting of Professor Shinnichiro Suzuki and Assistant Professor Masaki Nojiri at the Graduate School of Science, Osaka University, succeeded in clarifying the steric structure of the transient electron-transfer complex formed when copper-containing nitrite reductase reacts with an electron-donor protein for the first time in the world using the Macromolecular Assemblies Beamline, BL44XU (a contract beamline of the Institute for Protein Research, Osaka University) of SPring-8. This research achievement was published in the UK academic journal Nature on 5 November 2009. This study was supported by a Grant-in-Aid for Scientific Research and a Grant for Basic Science Research Projects from The Sumitomo Foundation. Publication: |
Background and achievement
The electron-transfer reaction between protein molecules is a process related to many reaction systems in living organisms, such as the photosynthesis system, the respiratory chain, and various metabolic systems, and it is necessary for the survival of living organisms. Similarly, in microorganisms, which carry out denitrification (NO3- → NO2- → NO → N2O → N2) and are intricately involved in the global nitrogen cycle, several reductases for catalyzing the stepwise reduction reactions starting from nitrate ions can effectively obtain the electrons necessary for their catalytic reactions from proteins, quinone molecules, or other molecules through intermolecular electron-transfer reactions. In general, transient complexes formed by molecules of electron-transfer partners are weakly bonded and have a short lifetime. This weak bonding is a physicochemical characteristic required for the efficient repetition of intermolecular electron-transfer reactions in living organisms. However, the number of reports on the high-resolution analysis of steric structures at the time of complex formation is limited because of the instability of the structures [there are only 10 examples in the protein databank (PDB)], and it is difficult to understand the mechanism underlying the electron-transfer reaction in detail.
In this research, Professor Shinnichiro Suzuki and Assistant Professor Masaki Nojiri of the Graduate School of Science, Osaka University, focused on the electron-transfer reaction between protein molecules induced by the reaction between nitrite reductase*1 (which produces nitrogen monoxide (NO), a precursor of nitrous oxide (N2O) that is attracting attention as a greenhouse gas, in the denitrification) and cytochrome c-551 as an electron-donor protein during denitrification. They determined the steric structure of the electron-transfer complex at a resolution of 1.7 Å by X-ray crystallography*2 using the Macromolecular Assemblies Beamline (BL44XU) at SPring-8.
In this complex, one cytochrome c-551 molecule binds to the nitrite reductase with a homotrimer structure composed of three subunits, each with a molecular weight of approximately 37 kDa (Fig. 1). The distance between the heme c*3 site of cytochrome c-551 and the type 1 copper site*4 of nitrite reductase, across which electron transfer occurs, is 10.5 Å; these sites are arranged to allow the redox reaction centers to be as close to each other as possible. At the interface formed by the two molecules, they interact, resulting in the exclusion of solvent water molecules from their surfaces; therefore, a hydrophobic environment is temporarily formed at the center of the interface (Fig. 2). Research and discussion on the electron-transfer reactions between protein molecules are expected to be markedly advanced by determining the types and positions of the amino-acid residues at the interactive interface and the position of the water molecules.
A research group at Leiden University (the Netherlands) analyzed the transient interaction between copper-containing nitrite reductase originating from a strain different from ours and pseudoazurin, an electron-donor protein, using nuclear magnetic resonance (NMR)*5 in 2008. Comparing the complex structure determined in this study with that suggested in their study, the interaction between nitrite reductase and cytochrome c-551 occurs at almost the same site on nitrite reductase as that between nitrite reductase and pseudoazurin, even though these electron-donor proteins, i.e., cytochrome c-551 and pseudoazurin, have completely different structures. This interaction is called a pseudospecific molecular recognition mechanism of the electron-transfer reaction between protein molecules. By determining the steric structure of the complex, the structural basis for pseudospecificity in the recognition mechanism of the electron-transfer partner of nitrite reductase was clarified. Moreover, in the analysis of the site-specific variant on the basis of the steric structure determined in this study, one of the amino acids playing a key role in the electron-transfer reaction was identified to be the methionine residue in the vicinity of type 1 copper (Fig. 3).
The electron-transfer system between nitrite reductase and cytochrome c-551 examined in this study is involved in the denitrification process, which plays an important role in balancing the global nitrogen cycle. As a result of this achievement, we have a more precise understanding of the molecular recognition mechanism of the two protein molecules as well as the intermolecular electron-transfer mechanism, both of which have been difficult to analyze by conventional methods. Accordingly, the development of devices to control the atomic nitrogen concentration at the global level is expected. As a result of recent global warming, nitrous oxide (N2O) generated in the denitrification process has been attracting attention as a greenhouse gas in addition to carbon dioxide, and it is no exaggeration to say that technology to control the global nitrogen cycle is eagerly expected as the next-generation technology following that to control carbon dioxide. The protein-protein interaction is a function associated with the lives of all organisms (cells) on Earth; and its strength, specificity, and binding-dissociation balance are controlled by a sophisticated mechanism in response to changes in the molecular environment. One reason why such a function can be realized is the flexibility unique to proteins. Therefore, we hope that this research achievement will contribute to the technological development of molecular switches using this flexibility.
<Figure>
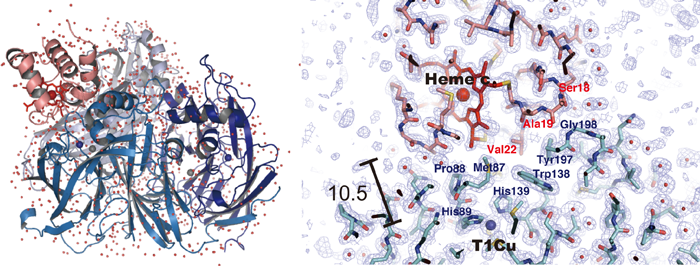
Fig. 1 Entire structure of complex of copper-containing nitrite
reductase and cytochrome c-551 (left) and site of interaction (right)
One molecule of cytochrome c-551 (pink ribbon) is placed on a nitrite
reductase molecule (homotrimer). The distance between the heme c
of the cytochrome c-551 molecule and the type 1 copper site of nitrite
reductase (heme c and T1Cu in the figure) may be as small as 10.5 Å.
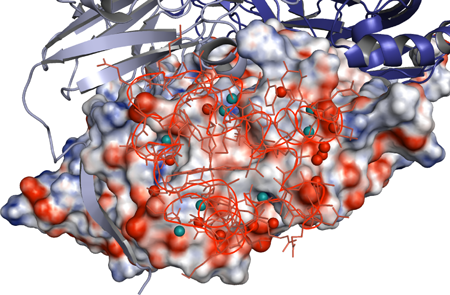
Fig. 2 Positions of water molecules at the interactive interface
Water molecules at the interactive interface are shown in light-blue
and red circles. The water molecules in light blue bond with both
cytochrome c-551 and nitrite reductase via hydrogen bonding, whereas
the water molecules in red bond with only one of them via hydrogen
bonding. At the center of the interface, no water molecules exist.
In the figure, the distribution of electric charge at the molecular
surface of the nitrite reductase subunit, with which cytochrome
c-551 interacts, is also shown (red, negative; blue, positive).
The cytochrome c-551 molecules are drawn using thin lines.
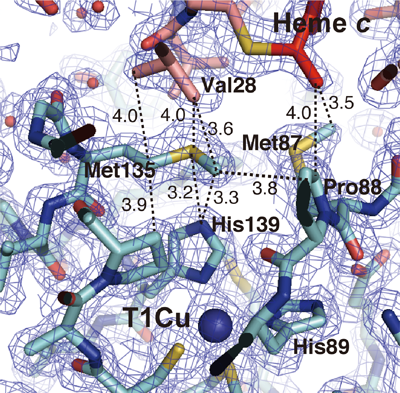
Fig. 3 Methionine residue, a key in the intermolecular electron-transfer reaction
When cytochrome c-551 binds with methionine 135 of nitrite reductase,
the methionine 135 is structurally fixed between the redox centers of two
molecules (heme c and type 1 copper). The rate constant of electron
transfer between the two molecules decreases to approximately one-tenth
when methionine is replaced with serine.
<Glossary>
*1 Nitrite reductase
Nitrite reductase is an enzyme that catalyzes the reduction of nitrite (ions). There are two types of nitrite reductase: dissimilatory and assimilatory. Dissimilatory nitrite reductase exists in denitrifying bacteria and is further classified into two types: those containing heme and those containing copper as a prosthetic molecule.
*2 X-ray crystallographic analysis
Crystals are made up of three-dimensionally ordered, regularly repeating patterns of atoms, molecules, or ions. X-ray crystallographic analysis is a method of analyzing the structure of a crystal by irradiating X-rays and examining their diffraction patterns and intensities. This technology is frequently used in the structural determination of proteins.
*3 Heme c
Heme c is an iron complex of porpherin c and is a prosthetic group, as is cytochrome c. Heme c is covalently bonded to the protein backbone by α-cysteinyl thioether linkages at positions 3 and 8.
*4 Type 1 copper site
A type 1 copper site generally refers to the binding site of mononuclear copper, which is retained inside a protein consisting of two histidine residues, one cystein residue, and one methionine residue. In rare cases, there are type 1 copper sites where the carbonyl oxygen, a principle component of the protein chain, is coordinated, glutamine instead of methionine is coordinated, and no methionine is coordinated. Owing to ligand-to-metal charge transfer (LMCT) from the sulfur atoms of the cysteine to copper atoms, the site has a strong blue color and is therefore also called blue copper.
*5 Nuclear magnetic resonance (NMR)
NMR is a technique for examining the structure of atoms and molecules. Approximately 20% of the steric structures of proteins have been determined by NMR. Dynamic structures can be determined because the structural determination of a material in a liquid is possible. Information on relatively weak molecular binding can also be obtained by NMR.
|
For more information, please contact: |
- Current article
- World's first determination of steric structure of transient electron-transfer complex formed when copper-containing nitrite reductase reacts with electron-donor protein during denitrification (Press Release)
 .
.