Discovery of New Mechanism of Drug Extrusion by Multidrug Efflux Proteins, a Cause of Multidrug-Resistant Bacterial Infection (Press Release)
- Release Date
- 28 Nov, 2011
- BL44XU (Macromolecular Assemblies)
Osaka University
A research group led by Ryosuke Nakashima (Assistant Professor) and Akihito Yamaguchi (Professor) of the Institute of Scientific and Industrial Research, Osaka University, has clarified the crystal structure of AcrB bound to drugs. AcrB is a multidrug efflux protein and the major cause of multidrug-resistant bacterial infection. This achievement is a groundbreaking discovery that leads to the clarification of the mechanism underlying the recognition and extrusion of drugs. Bacterial infection, which has been considered to have been conquered with the development of antibiotics, is coming back as a threat with the emergence of multidrug-resistant bacteria. In particular, there are still no effective clinical drugs against multidrug-resistant Pseudomonas aeruginosa. The major cause of multidrug resistance of Gram-negative bacteria, such as P. aeruginosa and Escherichia coli, is multidrug efflux proteins in these bacteria. Foreign objects, here, multidrugs, are supposedly unidentifiable by such proteins. The mechanism by which multidrug efflux proteins come to specifically recognize and extrude such supposedly unidentifiable drugs was an academic mystery. The scientists have studied the mechanism by examining the molecular structure of multidrug efflux proteins; in this research, they succeeded in determining for the first time the crystal structure of AcrB, which binds to high-molecular-mass drugs, using the SPring-8 beamline (BL44XU) for the crystal structure analysis of biological macromolecular assemblies. They found that the multidrug efflux proteins have multiple binding pockets that recognize various drugs, and they discovered for the first time the peristaltic pumping mechanism, by which high-molecular-mass drugs are transferred from one pocket to another. The structure of the molecular targets of inhibitors has been clarified, which has greatly advanced research on the development of molecularly targeted drugs used to overcome multidrug-resistant bacterial infection. This research was supported by the National Institute of Biomedical Innovation under the Program for Promoting Basic Research in the Field of Health and Medical Care and a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology. The results of this research were published online in Nature on 28 November 2011. Publication: |
<<Figures>>
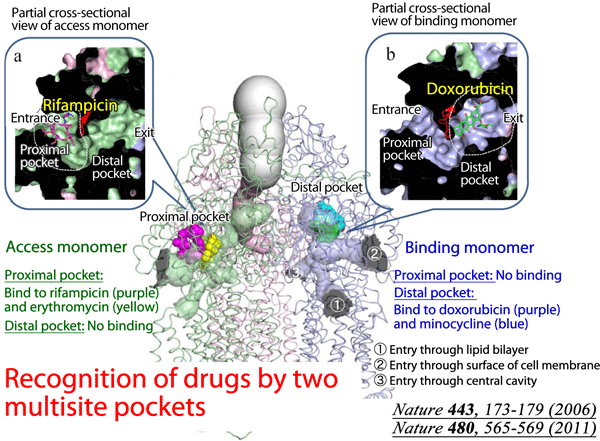
Fig. 1 In addition to the already clarified structure of the distal binding pocket, the proximal multisite drug-binding pocket was found in the structure of AcrB, which binds to high-molecular-mass drugs. These multisite drug-binding pockets with different substrate specificities markedly expand the range of substrates that AcrB can recognize.
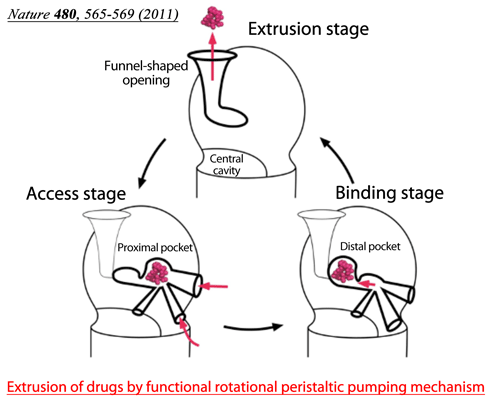
Fig. 2 Drugs enter the proximal pocket in the access stage, in which high-molecular-mass drugs are recognized, and are transferred to the distal pocket in the binding stage. Low-molecular-mass drugs are recognized by the distal pocket. The specificities for drug recognition are different between the two pockets.
|
For more information, please contact: |
- Current article
- Discovery of New Mechanism of Drug Extrusion by Multidrug Efflux Proteins, a Cause of Multidrug-Resistant Bacterial Infection (Press Release)