Mechanism of Maintaining Balance of Defense Responses to Stress (Press Release)
- Release Date
- 07 Nov, 2013
- BL32XU (RIKEN Targeted Proteins)
RIKEN
Key points
• Finding of Hfq forming a complex with catalase that decomposes poisonous hydrogen peroxide
• Discovery of a new mechanism that controls the function of Hfq and regulates the amount of stress-response proteins synthesized
• Providing important information on the Hfq-related protein regulatory mechanism, which had been unclarified
|
RIKEN (President, Ryoji Noyori) clarified part of the mechanism of maintaining the balance of biological defense responses to stress by experiments using Escherichia coli. This was achieved by the research group consisting of Koji Yonekura (associate chief scientist), Masahiro Watanabe [special researcher; currently, research scientist of the National Institute of Advanced Industrial Science and Technology (AIST)], and Yuko Kageyama (technical staff) of the Biostructural Mechanism Laboratory, Photon Science Research Division, and Saori Maki (research scientist) of the Biospecimen Platform Group, RIKEN SPring-8 Center (Director, Tetsuya Ishikawa). Living organisms synthesize the proteins necessary for defense responses in order to adapt to and survive various types of stress. In bacteria under stress, a protein called Hfq*1 binds to RNA to control the synthesis of stress-response proteins. Hfq and Hfq-like proteins are commonly seen across many species from bacteria to humans, all of which play important roles in binding to RNA and stabilizing its structure. However, little had been known about the mechanism of the control of Hfq-related protein synthesis. The research group found that Hfq and catalase*2, an enzyme that decomposes poisonous hydrogen peroxide, form a large complex when E. coli is grown under stress. The structural analysis of this complex at SPring-8*3 showed that catalase binds to the site that is involved in the binding between Hfq and RNA. Hfq cannot bind to RNA because this complex is formed when a large amount of catalase exists in bacteria, suppressing the excessive synthesis of stress-response proteins including catalase. These findings revealed that living organisms have a mechanism of maintaining the balance of defense responses to stress by controlling the functions of Hfq and regulating the amount of proteins synthesized. These findings will improve the understanding of many vital reactions related to Hfq and Hfq-like proteins. Moreover, it is expected that the structural information related to the mechanism of protein synthesis control will be applied to bioengineering. This research was partially supported by a Grant-in-Aid for Scientific Research Grant Number 20370064 from Japan Society for the Promotion of Science. Their achievements were published in the American scientific journal PLOS ONE on 6 November 2013. Publication: |
<<Figures>>
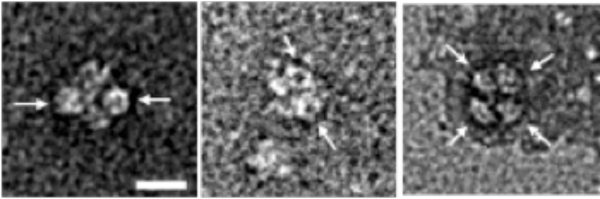
The ringlike structure of Hfq is indicated by arrows. The bar in the left image is 10 nm.
The bar is 100 μm.
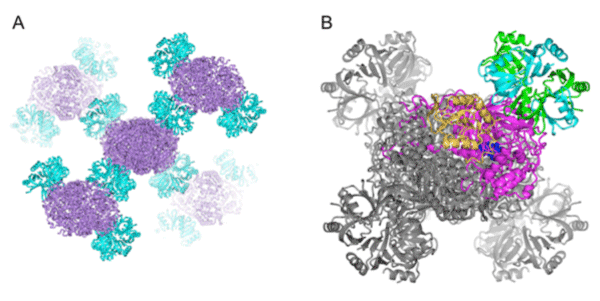
A: Molecular arrangement in crystals. Catalase and Hfq are indicated in purple and pale blue, respectively.
B: Enlarged view of Hfq-catalase complex. One of the Hfq rings, indicated in green and pale blue, binds to a catalase molecule, indicated in purple and yellow. Other molecular models are indicated in gray. The functional units of catalase and Hfq are a tetramer and a hexameric ring structure, respectively.
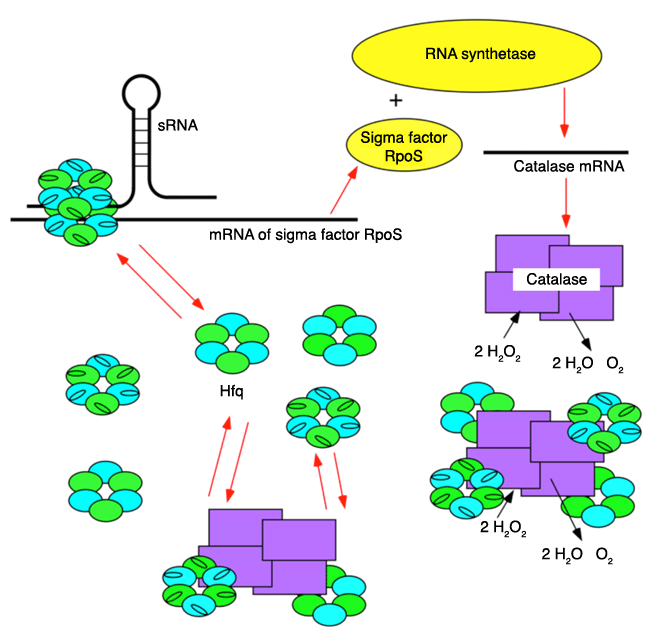
The synthesis of a sigma factor*6 RpoS, a cofactor of RNA synthetase*5, is stimulated by Hfq and sRNA*4 under stress. RpoS forms a complex with RNA synthetase and recognizes the catalase gene on DNA. The mRNA of the catalase gene is transcribed to synthesize catalase. When a large amount of catalase is accumulated inside cells, Hfq and catalase form a complex to inhibit the function of Hfq and to suppress the synthesis of RpoS. Thus, the production of catalase ceases and the cellular concentration of catalase is maintained at a constant level.
<<Glossary>>
*1 Hfq
An RNA-binding protein that forms a ringlike structure. Hfq and Hfq-like proteins are commonly seen across many species from bacteria to humans. Under stress, Hfq forms a hexameric ring in bacteria and, together with small RNA (sRNA), interacts with messenger RNA (mRNA) to regulate the stability and translation activity of mRNA. The translation reaction is thus controlled and regulated. The LSm protein, a close relative of Hfq in humans, forms a heptameric ring structure and is involved in mRNA editing. All of these proteins act as an RNA chaperone that stabilizes the RNA structure.
*2 Catalase
An enzyme commonly seen across many species from humans to various aerobic bacteria and that forms a tetramer. It rapidly decomposes poisonous hydrogen peroxide generated in cells by energy metabolism inside cells into oxygen and water.
*3 SPring-8
A facility that provides a wide spectrum of radiation light from far-infrared to visible, and soft X-rays up to hard X-rays. Research work in various fields, including nuclear science, nanotechnology, biotechnology, industrial use, and forensic investigation, is carried out at SPring-8. The crystallography of proteins performed at SPring-8 has also produced significant results.
*4 sRNA
A short RNA molecule that has no sequence information on proteins. A wide variety of sRNA molecules with various functions have been discovered. sRNA interacts with mRNA and regulates the translation of proteins.
*5 RNA synthetase
An enzyme that recognizes a DNA base sequence and synthesizes complementary mRNA. It is also called RNA polymerase.
*6 Sigma factor
A protein that determines the transcription initiation site in DNA. It determines the gene to be transcribed into mRNA by binding to RNA polymerase and recognizing a specific sequence in DNA. Living organisms have multiple sigma factors, and transcribe a group of genes that are appropriate to the environmental conditions by adjusting the proportion of the sigma factors inside cells in response to the changes in the environment. In E. coli in the stationary growth phase, the sigma factor RpoS is synthesized as the main factor for controlling the responses to various types of stress.
|
For more information, please contact: |
- Previous Article
- Chemical Synthesis of New Ball-Like Three-Dimensional Carbon Nanomolecules (Press Release)
- Current article
- Mechanism of Maintaining Balance of Defense Responses to Stress (Press Release)