Aiming at Development of New Drugs Against Influenza Virus
Influenza virus
Every winter there is concern about a possible pandemic of influenza. The symptoms of influenza include cough and fever, similar to those of the common cold. However, the common cold is caused by bacteria, whereas influenza is caused by viruses (Fig. 1).
Influenza viruses have eight RNA genes, and there are two types of spikelike protein on the surface of virus particles. Because of their very simple structure, influenza viruses cannot self-replicate. Instead, they invade human host cells and use the replication system of these cells skillfully.
The two types of spikelike protein on the virus surface are hemagglutinin (HA) and neuraminidase (NA). HA binds to glycoproteins on the surface of cells. Via this binding structure, viruses invade the cells (referred to as infection). On the other hand, NA plays a role in the separation/release of progeny viruses from the cells. The amino acids that constitute these two types of protein easily vary (referred to as mutation); 16 types of HA and 9 types of NA have been found thus far. Thus, there are as many types of influenza virus as there are combinations of these proteins (H1N1 - H16N9). For example, the most fundamental virus infectious for humans is H1N1. The avian influenza virus, the pandemic of which has recently been of concern, is H5N1. The pathogenicity also differs among different types of influenza virus. For example, the Spanish flu that spread in 1918 and the Hong Kong flu detected in 1968 were both highly virulent influenza viruses that claimed many lives (Fig. 2).
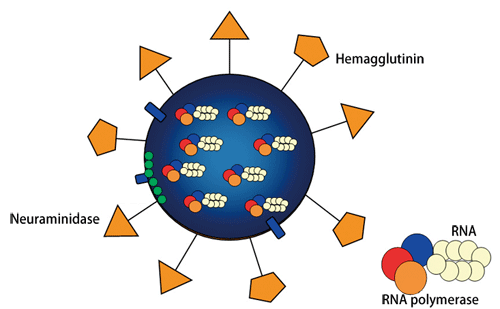
Fig. 1 Schematic of influenza virus structure
Many spikelike proteins are present on the surface. HA functions in viral invasion of a human host cell, whereas NA functions in viral release from the human host cell.
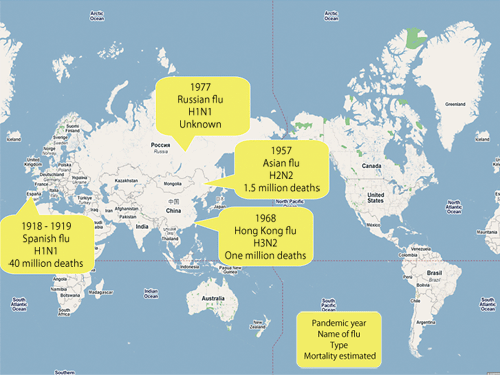
Fig. 2 History of pandemic influenza viruses
Difficulties in treating influenza
Current measures against influenza are prevention by vaccination and treatment with antiviral drugs such as Tamiflu®. Vaccines are prepared by predicting the type of influenza that is likely to spread each year. When a vaccinated person is infected with a virus, an immune reaction occurs immediately, thus preventing the virus from replicating in the body. However, vaccination has a disadvantage in that the vaccine is only effective against the corresponding type of influenza virus.
Tamiflu® is a drug that inhibits the release of viruses from infected cells. Unless a patient is administered Tamiflu® within 48 hours of infection, it has no effect on stopping the release of viruses. At any rate, there is no radical treatment for influenza, and the speed of recovery depends on the stamina of patients. Therefore, influenza is a threat for children and the elderly, who tend to have low stamina.
According to Dr. Sam-Yong Park, an associate professor at the Division of Protein Design, Department of Supramolecular Biology, Graduate School of Nanobioscience, Yokohama City University, avoiding contact with infected patients is still the best method of avoiding infection. He has analyzed the structure of an enzyme related to the replication of influenza viruses. In July 2008, he clarified the structure of an important section of a protein, which will lead to the development of new drugs.
Initial investigation of the enemy
Dr. Park considered types of protein to focus on during structural analysis to realize the development of new drugs, and concluded that he may obtain answers by focusing on the mechanism underlying the replication of influenza viruses.
Influenza viruses replicate their own genes and synthesize proteins in host cells (Fig. 3). New copies of viruses are formed from the thus-produced genes and proteins, and finally are released from the human host cells.
To carry out such replication, influenza viruses use 10 types of protein. One of them is RNA polymerase. This protein (an enzyme) is important for the replication of viral genes. RNA polymerase comprises the following three subunits: PA, PB1, and PB2 [Fig. 4 (left)]. When any of them is lacking, RNA polymerase loses its enzymatic function and viruses cannot replicate.
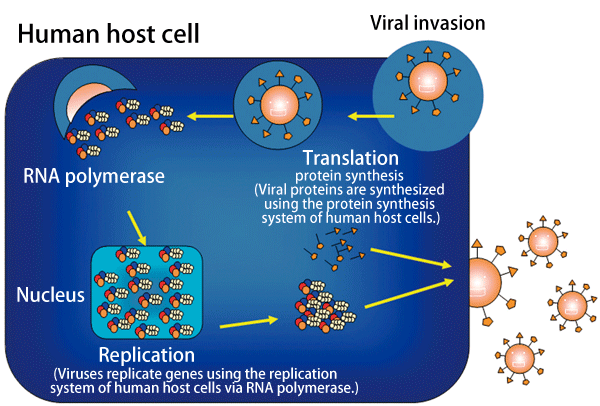
Fig. 3 Replication mechanism of influenza viruses
Influenza viruses invade host cells, produce genes by making use of the replication system of the host cells, and synthesize proteins.
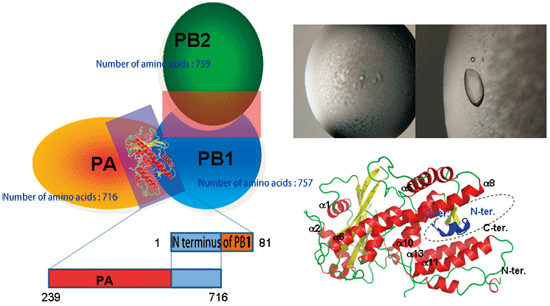
Fig. 4 Subunit structure of influenza RNA polymerase (right). Schematic of RNA polymerase subunits, PA, PB1, and PB2 (left).
The PA(239-716)/PB(1-81) complex was successfully crystallized (upper right). When crystallography was performed using the SPring-8 beamline BL41XU, 15 (blue) out of 81 amino acids at the N-terminus of PB1 are held in the keyhole structure (dotted circle) of the C-terminus of PA (lower right).
Structural analysis of RNA polymerase
“If we can determine the steric structure and understand how the three subunits bind to each other, we might be able to stop the function of RNA polymerase by artificially breaking the binding between these subunits.” To this end, Dr. Park decided to examine the binding structure between PA and PB1.
A considerable amount of proteins are required for the analysis of the protein structure. Because it is impossible to obtain a large amount of proteins from viruses, Dr. Park used coliform bacteria to synthesize proteins. This is a general method adopted when a large amount of proteins are needed. However, coliform bacteria were unable to synthesize stable PA and PB1 subunits.
The aim of their research at that time was to clarify the structure of the binding site between PA and PB1. Considering that there is no need to reproduce the entire structure, Dr. Park attempted to reproduce only the binding site by cleaving PA and PB1. However, it was unclear which parts of PA and PB1 bind to each other; therefore, various combinations of PA and PB1 subunits of different lengths were synthesized using coliform bacteria. After testing 200 or more combinations, it was found that a stable subunit can be obtained by combining the 239th - 716th sites of PA amino acids with the 1st - 81st sites of PB1 amino acids.
Next, the thus-obtained stable subunit was crystallized and analyzed by X-ray crystallography. Initially, they carried out experiments in a synchrotron radiation facility other than SPring-8, but the resolution was not satisfactory, and they could only determine the structure of the principal chain of an amino acid. Then, they used high-brilliance X-rays at the SPring-8 beamline BL41XU to carry out a detailed analysis of the steric structure [Fig. 4 (right)]. At the PA-PB1 binding site, the PB1 subunit was held in the pocket of the PA subunit, similarly to a key in a keyhole. According to Dr. Park, who was astonished at this result, the subunits of such large proteins are generally in contact with each other at the binding site, and it is rare that one subunit is held in another subunit at the binding site.
Furthermore, he carried out experiments by changing the amino acids corresponding to the PA keyhole and identified the amino acids important to the binding to PB1.
Potential for development of new drugs
RNA polymerase loses its function when any of the three subunits is lacking. Therefore, any substance that can inhibit the binding between the subunits of RNA polymerase can be effectively used for drugs against influenza. Several substances that inhibit the binding between PA and PB1 have already been identified.
The use of RNA polymerase to develop new drugs brings another great advantage. Compared with other proteins, RNA polymerase hardly mutates. That is, every influenza virus has common amino acid sequences. This is because, according to Dr. Park, RNA polymerase plays an important role in the replication of viruses and thereby hardly mutates. This means that drugs developed using RNA polymerase will be effective for all types of influenza virus.
Dr. Park has resolved a number of problems with his valuable ideas and has succeeded in clarifying the binding structure between PA and PB1. Application of this achievement to the development of new drugs will require much more effort and higher costs, for example, for examining many candidate materials for drugs and investigating their effects on humans. This cannot be carried out in a single laboratory. He says, “We would like to cooperate with other laboratories and pharmaceutical companies to realize the practical application of new drugs.” The winter that people need not worry about influenza may not be far off.
Column: 20 happy years in Japan in various environments
Dr. Park told me with a smile why he decided to attend a university in Japan saying, “Japan is close to my country.” He returned to Pusan, South Korea, at weekends when he was an undergraduate student at Osaka University. It took only 20 minutes for him to reach Itami Airport from Toyonaka campus of Osaka University by bicycle. From there the flight to Pusan took about one hour. He said, “I thought that I could go back to my country whenever I wanted, but I have found that 20 years have passed since I came to Japan.” This is because he was able to easily adapt to Japanese culture and food.
He looked back over a period of 20 years; he has studied at various laboratories, and particularly enjoyed four years from 1996 at SPring-8, he enjoyed being close to nature and very much enjoyed playing sports in an environment far from the city. “Now that my child(ren) has grown up, I cannot go back to my country,” he says. His candid personality becomes clear when he speaks.
 Dr. Park at Arhus, Denmark, where he attended an international conference
Dr. Park at Arhus, Denmark, where he attended an international conferenceInterview and original text by Akiko Ikeda (Sci-Tech Communications Incorporated)
This article was written after an interview with Dr. Sam-Yong Park at the Department of Supramolecular Biology, Graduate School of Nanobioscience, Yokohama City University.