Clarification of Nanostructure of Bacterial Flagellar Micropropeller that Switches Conformation (Press Release)
- Release Date
- 15 Mar, 2010
- Electron Microscope @ Osaka University
Osaka University
RIKEN
|
A joint research team consisting of researchers from Osaka University (Kiyokazu Washida, President) and RIKEN (Ryoji Noyori, President) analyzed the supermolecular filament structure of the flagellum, the swimming organ of a bacterial cell by cryomicroscopy and helical image analysis, and succeeded in clarifying the formation of the small bio-micropropeller and molecular switching mechanism. This was achieved through a joint research by Saori Maki, a research scientist of the Protein Crystallography Research Group, RIKEN SPring-8 Center (Tetsuya Ishikawa, Director); Koji Yonekura, an associate chief scientist of the Biostructural Mechanism Laboratory, RIKEN SPring-8 Center; and Keiichi Namba, a professor of the Graduate School of Frontier Biosciences, Osaka University. The achievements were published online in an American scientific journal, Nature Structural & Molecular Biology, on 14 March 2010 (London time). Publication: |
<Background of research>
Flagella, the locomotory organ of bacteria, are nanomachines composed of approximately 30 different proteins, and the structure of each component of a flagellum is surprisingly similar to that of an artificial electric motor. A flagellum has structures corresponding to a rotor, a stator, a reverse controller, a bearing, an adjustable joint, and a propeller, and the protein components of each structure have self-organizing capability (Fig. 1). A flagellar filament is a helical micropropeller formed by polymerization of 20,000-30,000 subunits of a protein, flagellin. The length of the flagellar filament is approximately tenfold the bacterial length, i.e., 10-15 μm. During swimming, the flagellar filament has a supercoiled assembly with a gentle curvature. However, when swimming toward a chemical substance such as a nutrient or escaping from a low-temperature aqueous environment, the rotation of the flagellar motor, which rotates at a high rate of approximately 20,000/min, reverses in about 1 ms. At this moment, the left-handed microscopic supercoil of the flagellar filament is temporarily reversed to the right-handed supercoil. Two different conformations, i.e., L-type and R-type filaments, of flagellin play a key role in this switching, which has remained unclarified thus far.
<Research contents and achievements>
The research group reported the R-type filament in 2003. Since then, the researchers promoted the technological development of cryomicroscopy and helical image analysis; this time, they succeeded in developing an atomic model of the L-type filament (Fig. 2). The molecular mechanism underlying the conformational switching of the flagellar filament induced by the structural difference between the L-type and R-type filaments was clarified. It was found that the close intermolecular interaction in the inner cylinder and flexible intermolecular interaction in the outer cylinder of the bicylindrical structure existing at the center of a flagellar filament underlie the morphological switching of the helical flagellar filament. The mechanism of control by a flexible bio-nanomachine was clarified (Fig. 3). Thus, one of the mechanisms controlling bacterial swimming, which has been unresolved since the 1960s, was revealed. We hope that this achievement will be useful for designing a propeller of a nanomotor as a nanotechnological application in the future.
<Future prospects>
There are various supermolecular complexes that form a filament structure in living organisms. However, the synthesis of crystals of these complexes is difficult and analysis of the molecular structure by high-resolution X-ray crystallography is extremely difficult. The present study is significantly important in terms of the possibility of developing a method of analyzing the natural functional structure of biological supermolecules, without requiring the synthesis of crystals.
<Figure>
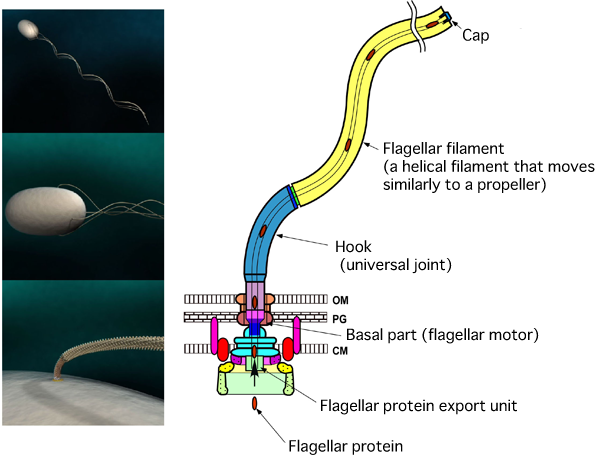
Salmonella and Bacillus coli swim by rotating bundles of flagella extending from the cell. The flagellum is a helical screw propeller with a helical pitch of 2.5 μm and a diameter of 0.5 μm. There are flagellar motors at the root of each flagellum. A flagellum consists of three parts, i.e., (1) a basal flagellar motor, (2) a hook as a universal joint, (3) a flagellar filament that moves similarly to a propeller (CM, cell membrane; PG, peptidoglycan layer; OM, outer membrane).
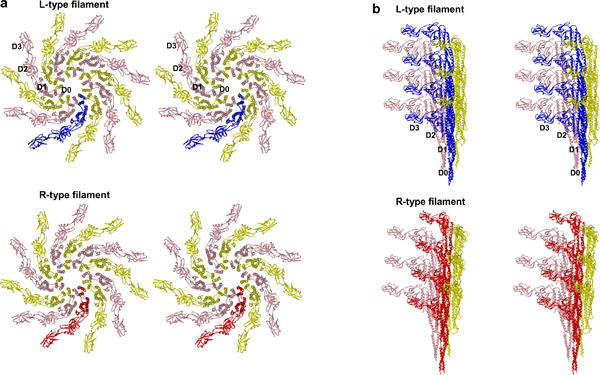
(a) Observed from the filament axis. The diameter of a flagellar filament is 240 Å. (b) Flagellin 9 observed from the side of a filament.
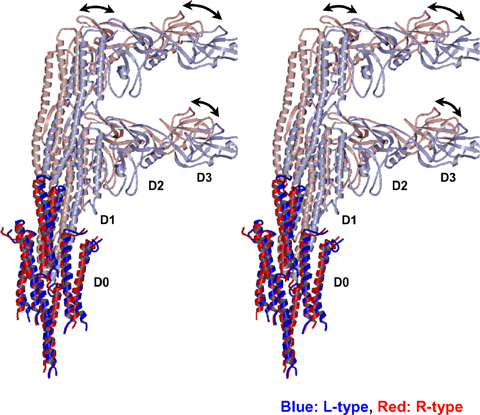
In the inner domains of the filament shown in solid red and blue, the difference between the L-type and R-type filaments is small; whereas in the outer domains shown in light red and blue, the two types of filament markedly differ at the points indicated by the arrow.
For more information, please contact: Dr. Koji YONEKURA (RIKEN) |
- Current article
- Clarification of Nanostructure of Bacterial Flagellar Micropropeller that Switches Conformation (Press Release)