Clarification of Mechanism of Specific Binding of DNA-Repair-Promoting Protein to Single-Stranded DNA (Press Release)
- Release Date
- 17 Mar, 2010
- BL26B2 (RIKEN Structural Genomics II)
- Success in crystallization and X-ray crystallographic analysis of RecJ protein, a DNA-repair-promoting protein from Thermus thermophilus -
RIKEN
|
Scientists at RIKEN (Ryoji Noyori, President) clarified the steric structure of RecJ protein, which degrades single-stranded DNA and is involved in the DNA repair mechanism*2 as a fundamental life phenomenon, using Thermus thermophilus HB8, an extremely thermophilic bacterium.*1 The bacteria thrives at 85°C and is considered to be an organism present at the beginning of life. They also clarified at the atomic level that the structure of the RecJ protein is responsible for its high affinity to single-stranded DNA. This was achieved by Taisuke Wakamatsu, a collaborative scientist, and Seiki Kuramitsu, group director, of the Synchrotron Radiation (SR) System Biology Research Group, RIKEN SPring-8 Center (Tetsuya Ishikawa, Director) under the Structural-Biological Whole Cell Project.*3 DNA carries the genetic information of living organisms; the rewriting of genetic information is a driving force of evolution, whereas at the same time, it involves a risk of cellular death and cancer. DNA is replicated with every cell division; however, DNA replication errors may sometimes occur. In addition, DNA is constantly damaged by extrinsic factors, such as ultraviolet rays present in sunlight, and there is a possibility that rewriting of the DNA sequence may occur. Living organisms, however, have a DNA repair mechanism by which damaged DNA can be repaired, and the mechanism is considered to be basically the same for bacteria and humans. It has already been clarified that single-stranded DNA degradation proteins, such as the RecJ protein, play an important role in removing DNA replication error sites. However, the detailed steric structure of the RecJ protein and the reason why it specifically degrades single-stranded DNA have remained unclear. The research group carried out X-ray crystallographic analysis at SPring-8 to clarify the steric structure of the RecJ protein of Thermus thermophilus HB8. The group found that the RecJ protein forms an O-shaped structure that wraps single-stranded DNA and that it has a typical structure enabling nucleic acid binding. The structure is unique and has not been reported thus far for single-stranded DNA degradation proteins. Because the steric structure of the RecJ protein was determined, it is expected that the detailed functions of other single-stranded DNA degradation proteins with an amino acid sequence similar to that of the RecJ protein will be clarified, which may significantly contribute to the elucidation of the DNA repair mechanism of living organisms. The achievements were published in an American scientific journal, Journal of Biological Chemistry, on 26 March 2010. Publication: |
<Figure>
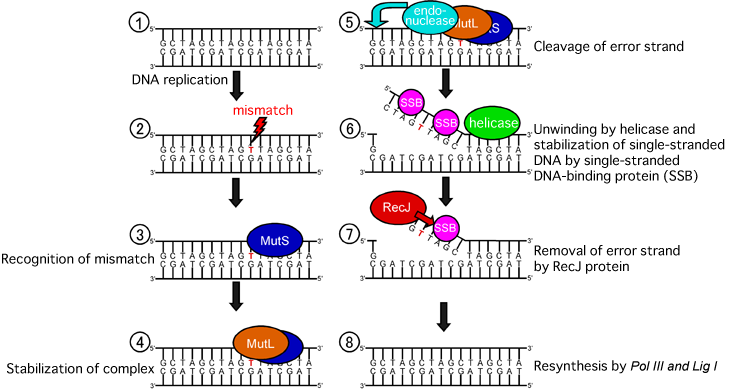
During DNA replication ( ), if a mismatch (
), if a mismatch ( ; Originally, C instead of T should bind to G) occurs, the MutS protein finds the mismatch and binds to the DNA (
; Originally, C instead of T should bind to G) occurs, the MutS protein finds the mismatch and binds to the DNA ( ). At the same position, the MutL protein binds (
). At the same position, the MutL protein binds ( ), and furthermore, an endonuclease, a DNA degradation protein, cleaves the single-stranded DNA with an error (
), and furthermore, an endonuclease, a DNA degradation protein, cleaves the single-stranded DNA with an error ( ). A protein helicase separates the single-stranded DNA and prevents a single-stranded DNA-binding protein (SSB) from binding to the single-stranded DNA to form a double-stranded DNA again (
). A protein helicase separates the single-stranded DNA and prevents a single-stranded DNA-binding protein (SSB) from binding to the single-stranded DNA to form a double-stranded DNA again ( ). SSB binds to the RecJ protein to degrade the error strand (
). SSB binds to the RecJ protein to degrade the error strand ( ). Finally, a double-stranded DNA is resynthesized by polymerase III (Pol III) and DNA ligase I protein (Lig I) (
). Finally, a double-stranded DNA is resynthesized by polymerase III (Pol III) and DNA ligase I protein (Lig I) ( ).
).
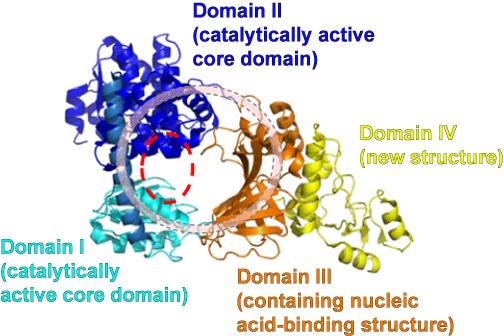
Fig. 2 Overall structure of RecJ protein from Thermus thermophilus
The RecJ protein forms an O-shaped structure by connecting the four domains, namely, domains I and II constituting a catalytically active core domain, domain III containing an oligonucleotide/oligosaccharide binding fold, which is unique to DNA-binding proteins, and domain IV, the structure of which is newly clarified and the function of which is still unknown, in a ring shape. The catalytically active region is indicated by a dotted red line.
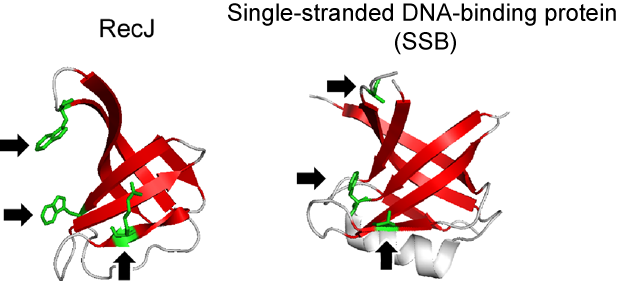
Fig. 3 Oligonucleotide/oligosaccharide binding fold of RecJ protein
and already-known single-stranded DNA-binding protein (SSB)
In domain III of the RecJ protein, amino acid residues (green) that interact with single-stranded DNA exist, similar to SSB. The structures of the regions involved in single-stranded DNA binding (red ribbons) are similar for both the RecV protein and SSB.
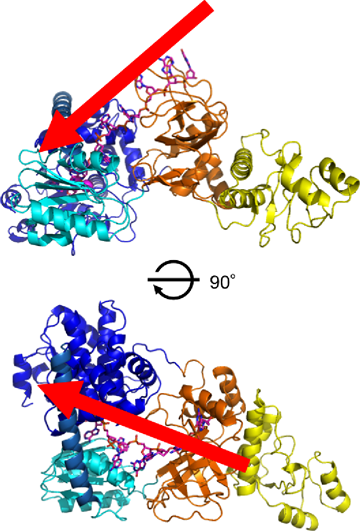
Pink indicates the model single-stranded DNA. It is expected that the single-stranded DNA advances in the direction indicated by the red arrow (from the DNA-binding structure in domain III to the core domain consisting of domains I and II) and degrades at the catalytically active region. The lower figure is obtained by rotating the upper figure back and forth by 90°.
<Glossary>
*1 Thermus thermophilus HB8
Thermus thermophilus HB8 is a bacterium that can thrive in an extreme environmental condition of 85°C and was isolated from Mine Hot Spring located on the Izu Peninsula in Shizuoka Prefecture. It is considered that bacteria living in hot water (thermophiles) are the common ancestors of all living organisms and that the fundamental characteristics of a protobiont are observed in these bacteria. Therefore, understanding life phenomena occurring in one thermophile may clarify the fundamental characteristics of all living organisms, including humans; it will also lead to answering the essential question of "what is life."
*2 DNA repair mechanism
DNA is constantly damaged by intrinsic factors, such as DNA replication errors and active oxygen species, and extrinsic factors, such as ultraviolet and ionizing radiation. However, living organisms have various mechanisms of repairing DNA to its original state, and many DNA-repair promoting proteins are involved in these mechanisms. The RecJ protein, the steric structure of which was clarified in this study, is considered to have functions such as DNA mismatch repair, base excision repair, and homogeneous recombination repair.
*3 Structural-Biological Whole Cell Project
This is a project aimed at establishing an academic foundation to systematically understand all the life phenomena observed in one cell on the basis of the structures and functions of DNA, proteins, carbohydrates, lipids, and other low-molecular-weight materials using Thermus thermophilus HB8 as a model representing all the living organisms on Earth (model organism). It is assumed that the project will proceed following the four steps listed below. At SPring-8, research related to imaging is carried out.
Step 1: Analysis of the steric structure of a whole cell, which is composed of molecules, such as proteins
Step 2: Analysis of the function of a whole cell, which is composed of molecules, such as proteins
Step 3: Analysis of various systems in a cell (network of multiple molecules)
Step 4: Simulation of the whole cell
For more information, please contact: |
- Previous Article
- Clarification of Nanostructure of Bacterial Flagellar Micropropeller that Switches Conformation (Press Release)
- Current article
- Clarification of Mechanism of Specific Binding of DNA-Repair-Promoting Protein to Single-Stranded DNA (Press Release)