Clarifying Steric Structure of Enzyme That Maintains Genomic Methylation Patterns - Leading to development of medicines that control cancer cells (Press Release)
- Release Date
- 26 Apr, 2011
- BL44XU (Macromolecular Assemblies)
Osaka University
|
Scientists of the laboratories led by Professor Shoji Tajima and Professor Atsushi Nakagawa of the Institute for Protein Research, Osaka University, have clarified the steric structure of an enzyme that transmits genomic methylation patterns to next-generation cells by X-ray crystallography, using the Macromolecular Assemblies Beamline (BL44XU) at SPring-8. Genomic methylation patterns are characteristically different among various tissues and are considered to be most important for the control of genetic information during epigenesis, which is studied in epigenetics.*1 When DNA is replicated, genomic methylation patterns are accurately transmitted to next-generation cells. This transmission is assisted by an enzyme called DNA methyltransferase 1 (Dnmt1). Cells cannot survive if methylation patterns are not maintained. However, how Dnmt1 maintains methylation patterns has not been fully understood. In this study, the steric structure of Dnmt1 was determined, and a clue to how methylation patterns are maintained was obtained, clarifying part of this complicated maintenance mechanism. The achievements of this research will lead to the development of medicines capable of controlling cancer cells that unlimitedly proliferate. The results of this study were published online as a paper entitled, “Structural insight into maintenance of methylation by mouse Dnmt1” (PNASMS#2010-19629RR) in the Proceedings of the National Academy of Sciences of the United States of America on 25 April 2011. Publication: |
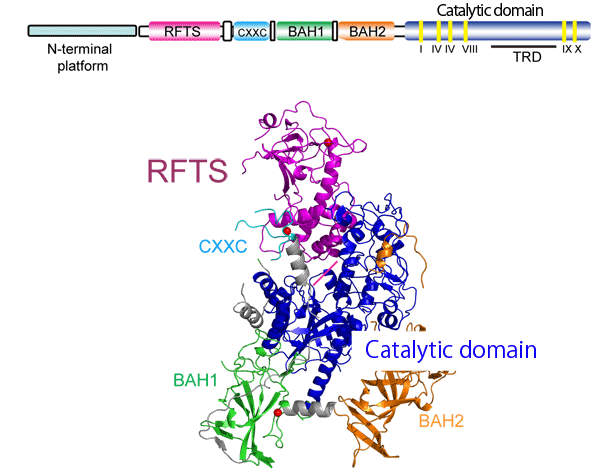
<Figure>
Fig. 1 Crystal structure of mouse Dnmt1 (2.75 Å)
The Dnmt1 structure is as follows: 1. the replication focus-targeting sequence (RFTS, shown in pink), which localizes Dnmt1 to the DNA replication starting point from the N-terminus, 2. CXXC (light blue), and 3. two bromo-adjacent homology (BAH) domains (green and orange). These components are arranged around a catalytic domain (blue) of the C-terminus, forming its domain structure. Unexpectedly, RFTS is positioned such that it pierces the catalytic pocket, forming a structure to which hemimethylated DNA,*2 a methyl-group receptor substrate, cannot bind. On the basis of the comparison of the activity of Dnmt1 with that of Dnmt1 from which RFTS was knocked out, it was speculated that Dnmt1 has a mechanism of releasing RFTS from its catalytic pocket.
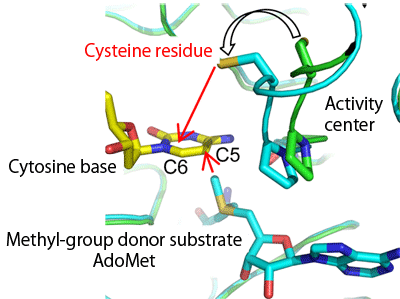
From the similarity to bacterial methyltransferase, the 1229th cysteine residue at the catalytic center of Dnmt1 is considered to form a covalent bond with C6 of the target cytosine base as a reaction intermediate. The analysis of the structure of a complex with AdoMet, a methyl-group donor substrate, revealed that the side chain of the cysteine residue is flipped towards the cytosine base of DNA, which is expected to be bound to the cystein residue. Such flipping of the side chain of the cysteine residue towards C6 of the cytosine base is not observed for the complex of bacterial methyltransferase with AdoMet, and is thereby unique to Dnmt1.
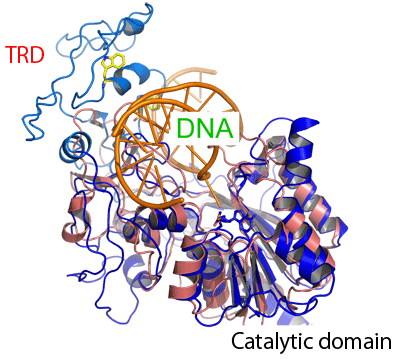
When hemimethylated DNA is superimposed to the catalytic local structure (blue) of Dnmt1 on the basis of the structure of a complex of bacterial methyltransferase and DNA, it is speculated that a target recognition domain (TRD), which recognizes a target sequence in bacterial DNA methyltransferase, is positioned such that it covered the methylated cytosine on the opposite strand of the target methylated cytosine. This TRD is considered to be important in recognizing hemimethylated DNA.
<Glossary>
*1 Epigenetics
There is no doubt that DNA base sequences carry information on proteins constituting the human body; however, exactly identical humans cannot be produced even if individuals have the same base DNA sequences, as in the case of monozygotic twins. The same DNA base sequences may show different expression patterns. The mechanisms underlying the transmission of genetic information to next-generation cells caused by gene expression independent of the underlying DNA base sequences are studied in the field of epigenetics. DNA modification and the control of genome-folded structures are also the focus of interest in epigenetics.
*2 Hemimethylated DNA
One strand of double-stranded DNA is methylated. Daughter DNA strands newly generated after DNA replication are only partially methylated, i.e., they are hemimethylated.
|
For more information, please contact: |
- Previous Article
- Magma Ocean May Have Extended across the Bottom of the Mantle in Primitive Earth (Press Release)
- Current article
- Clarifying Steric Structure of Enzyme That Maintains Genomic Methylation Patterns - Leading to development of medicines that control cancer cells (Press Release)