Elucidation of the Mechanism Creating the Small RNA that Inhibits Genome Mutation (Press Release)
- Release Date
- 15 Oct, 2012
- BL32XU (RIKEN Targeted Proteins)
Japan Science and Technology Agency (JST)
The University of Tokyo, Graduate School of Science
Main points
• Transposon (a mobile gene) gives rise to genome mutation, but a certain small RNA is capable of inhibiting it.
• Creating the small RNA requires an RNA slicing enzyme. This research successfully identified it.
• The research results may hold promise toward the elucidation of the pathogenic mechanism of infertility and the development of reproductive medicine.
|
The research group, at the University of Tokyo, Graduate School of Science, revealed a part of the molecular machinery in which a small RNA (ribonucleic acid) - responsible for protecting a genome (entire gene) - is created. The research was conducted as part of JST Mission-oriented Basic Research, and the group consisted of, among others, Prof. Osamu Nureki, Prof. Mikiko Shiomi, Assistant Prof. Hiroshi Nishimasu (special-appointment) and Hirotsugu Ishizu. Animal genomes include a type of mobile gene, called a transposon,*1 whose transfer in many cases is known to cause genome damage, inducing a variety of diseases. Living organisms have a built-in mechanism to regulate transposon expression and suppress transfer activity. In reproductive cells that are responsible for inheriting genetic information in an accurate fashion, a small RNA (piRNA*2) undertakes the role of protecting the genome from being damaged. Our basic understanding is that this RNA is composed of a long single-stranded RNA, but the mechanism by which it is created has been unknown. The research group focused on the Zucchini (Zuc) protein,*3 which is a class among the many proteins known to have a hand in piRNA production of drosophila and mice. Firstly, the crystal structure of Zuc protein was analyzed using the ultra-high intensity microbeam available at SPring-8 (BL32XU line), which revealed that the protein has an optimized structure for slicing single-stranded RNAs. Subsequent biochemical analysis clearly indicated that the Zuc protein features enzyme activity capable of slicing a single-stranded RNA, supporting the results from X-ray crystal structure analysis. Additionally, cell biological analysis made it clear that the RNA slicing activity of the Zuc protein constitutes an integral part in producing piRNA and inhibiting transposon expression. The true identity of the enzyme that slices a single-stranded RNA, required for producing piRNA, has long been in question, until this research indicated that the Zuc protein assumes the role. Experimental evidence obtained using drosophila and mice has proved that the mutation in the Zuc gene leads to infertility, and that animals (including humans) also possess Zuc proteins. Toward the future, the results of this research may hold significant promise for further elucidation of the pathogenic mechanisms of infertility in humans and animals. The findings reported here were obtained through collaborative research with Prof. Haruhiko Shiomi (Keio University, School of Medicine) and Prof. Junken Aoki (Tohoku University, Graduate School of Pharmaceutical Science), and published in the online version of Nature (a UK science journal) on the 14th of October, 2012 (BST 18:00). Publication: |
Background and history of the research
Eukaryote’s genome has a mobile gene called a transposon. As a transition of a transposon from one genome position to another can result in a change in genetic information, to suppress the expression and transition of transposons is of great importance, especially for reproductive cells that are responsible for the accurate inheritance of genetic information to the next generation. Living organisms have a built-in mechanism to regulate transposon expression and suppress transition, and the occurrence of irregularities in this mechanism is known to trigger abnormality in the formation of sperms and eggs, leading to infertility.
All research up to the present have demonstrated that a type of small RNA (consisting of a base of 30 or so) expressed in reproductive cells assumes the role of suppressing transposon expression, thus protecting the genes in reproductive cells from being modified. It is known as piRNA (PIWI-interacting RNA) and functions as, so to speak, a quality control agent. The conventional idea of piRNA generation is that a long single-stranded RNA, created in a specific region of genome, is sliced into smaller ones through the operation of a certain type of enzymes. It has also been known that several proteins, including the Zucchini (Zuc) protein, have a hand in producing piRNA, but the mechanism through which a piRNA is produced has remained unexplained.
Details of the research
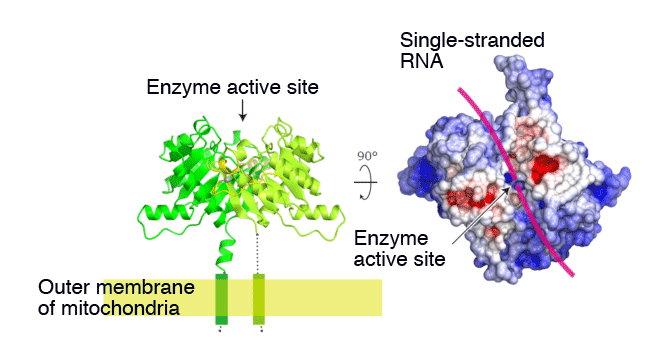
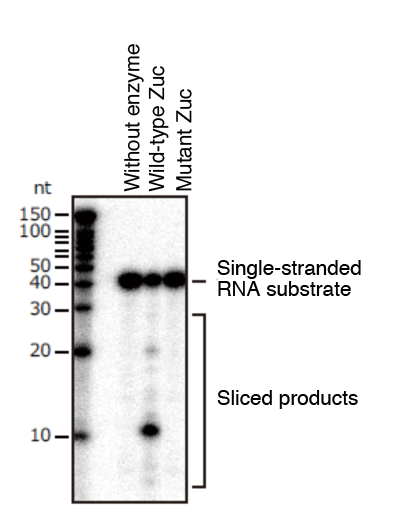
The research group focused on structural and functional details of the Zuc protein: a type of protein believed to be involved in the piRNA production of drosophila. The first step was to obtain the drosophila-derived Zuc proteins in quantity and to purify to them to a very high degree for crystallization, which we succeeded in doing. The crystallized sample was then served for X-ray structural analysis using the ultra high-intensity microbeam (BL32XU line) available at SPring-8, leading to the elucidation of the Zuc protein crystal structure from the mass of high-resolution X-ray diffraction data. The analysis made it clear that the Zuc protein constitutes a dimer - from two monomers, each made of a single polypeptide chain - and that the interface between the monomers has enzyme active sites with optimum configuration for slicing a single-stranded RNA (Fig. 1). The analysis also indicated that the Zuc protein has a favorable shape for localized functioning on the outer membrane of mitochondria, in good agreement with the conventional GFP fluorescent imaging data. Further, examinations of enzymic function of the purified Zuc protein showed that one of its actions is to slice a single-stranded RNA (Fig. 2). As expected from the configuration of the enzyme active site, it did not slice a double-stranded RNA. For further information, we examined the effect of modified enzyme active sites on piRNA production and the amount of transposon expression: we prepared a mutant Zuc protein by modifying its enzyme active site and introduced it to the reproductive cells of drosophila. The examination proved that the RNA slicing function on the part of Zuc proteins is integral both for piRNA production and suppressing transposon expression.
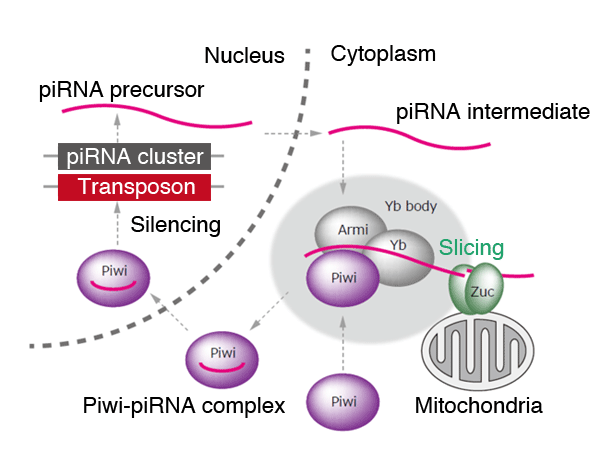
As described above, the functionality of the Zuc protein expected from X-ray crystal structure analysis showed good agreement with the evidence obtained from biochemical and cell biological analysis. The knowledge gained from these experiments strongly indicates that the Zuc protein assumes the role of an RNA-slicing enzyme, which constitutes an integral part in the piRNA producing mechanism (Fig. 3).
Future developments
Although all the research up to the present has proved that certain proteins - including the Zuc protein - play an important role in piRNA production, the specific functions of each protein have been largely unknown. As the function of the Zuc protein became identified by this research, we were able to unravel a part of the molecular mechanism for piRNA production. From here on, the whole picture of the transposon suppression mechanism is expected to be pieced together by identifying other proteins’ functions in detail. Based on the evidence gained from the research using drosophila and mice, it is known that anomalies in the proteins involved in the suppression of transposon-triggered genetic mutation can lead to infertility. Further advances in similar studies on transposon suppression in humans, especially those on the proteins controlling it, are expected to pave the way to the elucidation of the pathogenic mechanism of human infertility, and toward the development of fertility-treatment methods./p>
Additional statement
A part of the results reported here was obtained from a project conducted as a Targeted Proteins Research Program (Ministry of Education, Culture, Sports, Science and Technology) (head researcher: Dr. Osamu Nureki, member of the research project: Dr. Haruhiko Shiomi) supported under the auspices of Grants-in-Aid for Scientific Research.
<<Figures>>
(Left): The figure represents a ribbon model. Zuc forms a dimer, with an interface containing enzyme active sites. Two monomers are shown in green and greenish yellow respectively. The zinc-binding domain, involved in binding with RNA, is shown in yellow, and the grey balls indicate zinc ions. Zuc is considered to bind with the outer membrane of mitochondria by way of the transmembrane helix located at the N-terminal.
(Right): The figure represents a molecular surface model. The blue surface indicates that it is positively charged, and the red surface negatively charged. The substrate (a single-stranded RNA) is considered to bind with, and then be sliced from the groove in the enzyme active site. The single-stranded RNA is schematically shown as a magenta line. The groove width in the enzyme active site is so small that it possibly excludes combinations with a double-stranded RNA.
A purified Zuc protein and radioisotope labeled synthetic RNA were mixed for reaction, and then the mixture was served for analysis using polyacrylamide gel electrophoresis. In contrast to the wild type Zuc that severed single-stranded RNA, the Zuc with mutations in enzyme active sites did not sever the single-stranded RNA.
A piRNA precursor is transcribed from a genome region called a piRNA cluster. The piRNA precursor is sliced by some sort of RNase, producing a piRNA intermediate with a base length of several hundreds. The piRNA intermediate, in turn, is sliced by Zuc, producing a piRNA with a base length of about 30. In a granule-like structure called a Yb body, the piRNA is incorporated into a Piwi protein*4 by the operation of Armi proteins and Yb proteins. Finally, the Piwii-piRNA complex transitions into a nucleus, resulting in the suppression of transposon expression.
<<Glossary>>
*1 Transposon
Transposable elements present in the genome of eukaryotes, also called a mobile genetic element. Displacement of a transposon poses the possibility of disrupting genetic information, but it also allows a view that it has contributed to biological evolution.
*2 piRNA
Piwi-interacting RNA (piRNA) has a base length around 30 and is expressed in reproduction cells. piRNA belongs to a class of RNA that do not function as a blueprint of proteins (non-coding RNA). Many of them have an arrangement that complements a transposon, and, by binding with a PIWI protein, exerts an effect of hindering transposon expression.
*3 Zucchini (Zuc) protein
The Zucchini gene was originally identified, during a generic screening of drosophila, as the gene responsible for female sterility. Later studies have shown that the Zucchini protein has a certain hand in piRNA production, but the details of its function had remained elusive. The name Zucchini came from the fact that the eggs of a drosophila with mutation in the gene take an oblong shape (just like the vegetable Zucchini).
*4 PIWI protein
The protein that has the function of suppressing transposon expression through binding with piRNA. An animal has multiple PIWI proteins, for example, a drosophila has three (Piwi, Aub, and Ago3). The Piwi gene was originally identified, through genetic screening of the drosophila, as an essential gene required for the formation and maintenance of reproductive stem cells. The gene is tagged as Piwi (P-element induced wimpy testis) because its mutation can trigger hypogonadism.
|
For more information, please contact:
Prof. Mikiko Siomi (The University of Tokyo, Graduate School of Science) |
- Current article
- Elucidation of the Mechanism Creating the Small RNA that Inhibits Genome Mutation (Press Release)