Seeing Origin of Respiratory Enzyme from the Viewpoint of Proton Pathway (Press Release)
- Release Date
- 23 Jan, 2012
- BL41XU (Structural Biology I)
- BL44B2 (RIKEN Materials Science)
RIKEN
Key research achievements
·Clarification of crystal structure of quinol-dependent nitric oxide reductase (qNOR) related to anaerobic respiration
·Discovery of a structure corresponding to original model of proton pump of aerobic respiratory enzymes in an anaerobic respiratory enzyme
·Provision of a direction for designing artificial molecules by imitating molecular evolution
|
Scientists of RIKEN (President, Ryoji Noyori) analyzed the crystal structure of quinol-dependent nitric oxide reductase (qNOR)*1 produced by a thermophilic bacterium that performs anaerobic respiration, and discovered a structure corresponding to the original model of a proton pump*2 of aerobic respiratory enzymes. This discovery clarified part of the course of molecular evolution that respiratory enzymes had undergone over a period of several billion years. This was achieved by a research group led by Yoshitsugu Shiro, the chief scientist of the Biometal Science Laboratory of RIKEN SPring-8 Center (Director, Tetsuya Ishikawa), and Yuji Sugita, the associate chief scientist of the Theoretical Biochemistry Laboratory of RIKEN Advanced Science Institute (Director, Kohei Tamao). Many living organisms on earth including humans take oxygen into their body to survive. The oxygen is used as fuel for the synthesis of adenosine triphosphate (ATP),*3 which is utilized as energy in living bodies. Respiration using oxygen is referred to as aerobic respiration. In contrast, anaerobic respiration, in which compounds of nitrogen and sulfur instead of oxygen are used to synthesize ATP, takes place in microorganisms living in environments devoid of oxygen, such as the deep sea, soil, and hydrothermal areas. They are considered to be descendants of microorganisms that lived more than three billion years ago. cytochrome c oxidase (COX)*4 on the cell membrane plays a key role in aerobic respiration. COX pumps protons (H+)*5 from the inside to the outside of cells to generate a gradient in proton concentration across the cell membrane, allowing ATP synthase to efficiently function. In anaerobic respiration, the main anaerobic respiratory enzyme is nitric oxide reductase (NOR),*1 which has no proton pumping function, making ATP synthesis less efficient than that in aerobic respiration. Therefore, the fact that respiratory enzymes acquired the highly advanced function of proton pumping during the course of their evolution is considered to underlie the evolution of living organisms. Using a beamline at SPring-8,*6 the research group discovered a pathway through which protons necessary for the reduction of NO are transferred from the inside of cells to active sites by determining the crystal structure of qNOR from a thermophilic bacterium that performs anaerobic respiration. Although qNOR has no proton pumping function, its pathway was located close to the position of the proton pump of COX. A structure considered to be the original model of the proton pump was observed, for the first time in the world, in an anaerobic respiratory enzyme, which was previously considered to have no proton pump. This achievement clarified part of the course of molecular evolution that respiratory enzymes underwent over a period of several billion years, which is a discovery unveiling the origin of living organisms that perform aerobic respiration. The results of this research were published online in the scientific journal Nature Structural and Molecular Biology on 22 January 2012 prior to the publication in the printed version. Publication: |
<Figures>
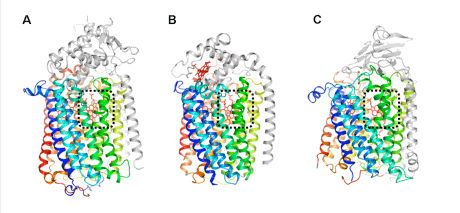
The figure shows the overall structures of (A) qNOR, (B) cNOR, and (C) NOR. The enzymatic core region shown in color has a similar structure among the three respiratory enzymes. Each enzymatic active site denoted by a black dotted line contains a heme molecule shown as a red stick, which is related to enzymatic activity and electron transfer.
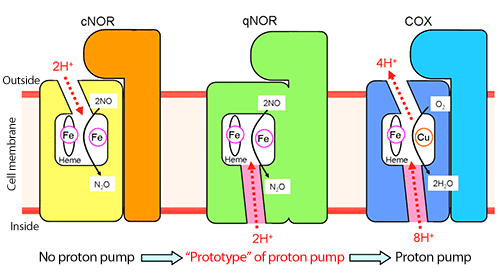
The figure shows the schematics of the proton transfer pathway for NO reduction in cNOR and qNOR and the proton pumping pathway in COX. The direction of proton transfer is denoted by red dotted arrows. As explained in established theories, a proton transfer pathway from the outside of the cell to the active site for NO reduction was observed in cNOR. In contrast, no proton transfer pathway from the outside of the cell was observed in qNOR; however, a proton transfer pathway from the inside of the cell to the active site was identified. In COX, protons are pumped from the inside to the outside of the cell through heme molecules in the active site. The position of the proton transfer pathway identified in qNOR in this study (shown in pink) is similar to that of the proton pumping pathway from the inside of the cell to the active site in COX.
<Glossary>
*1 Nitric oxide reductase (NOR, qNOR, cNOR)
NOR is a respiratory enzyme that reduces nitric oxide (NO) to synthesize nitrous oxide (N2O) (2NO + 2H+ + 2e- → N2O + H2O). There are roughly two types of NOR with different amino-acid sequences and electron donors: NOR using quinol in the cell membrane as the electron donor is called quinol-dependent NOR (qNOR), and NOR using cytochrome c, an electron transfer protein outside of cells, as the electron donor is called cytochrome c-dependent NOR (cNOR).
*2 Proton pump
The proton pump refers to a reaction system in living bodies in which protons are actively transferred from the inside to the outside of a cell using optical and chemical energies to generate a gradient in proton concentration across the cell membrane. The gradient in proton concentration is necessary for synthesizing ATP, which is utilized as energy in living bodies.
*3 Adenosine triphosphate (ATP)
ATP is an energy source commonly used by living organisms on earth. ATP is required in the reactions within the living body that require energy.
*4 cytochrome c oxidase (COX)
COX is the most important enzyme in aerobic respiration and reduces oxygen molecules using electrons and hydrogen ions (O2 + 4H+ + 4e- → 2H2O). The driving force required in the synthesis of ATP is generated via this reaction.
*5 Proton (H+)
A proton refers to a hydrogen ion. This is a monovalent cation, obtained when a hydrogen atom loses an electron (e-). Protons and electrons are involved in various chemical reactions that accompany redox reactions.
*6 SPring-8
SPring-8 is the world’s largest synchrotron radiation facility located in Sayo-cho, Sayo-gun, Hyogo Prefecture and is capable of generating intense X-rays. BL41XU and BL44B2 (the latter is currently used as a materials science beamline for powder diffraction using high-energy synchrotron radiation) used in the X-ray diffraction experiments in this study enable the acquisition of high-resolution data from laboratory X-ray generators for minute crystals that are difficult to structurally analyze.
|
For more information, please contact: |
- Previous Article
- Clarification of Metallic Transition of Iron(II) Oxide (FeO) with Rock-Salt-Type Structure Induced by High Pressure and High Temperature (Press Release)
- Current article
- Seeing Origin of Respiratory Enzyme from the Viewpoint of Proton Pathway (Press Release)