Exploration of Origin of Acquired Immunity (Press Release)
- Release Date
- 11 Oct, 2013
RIKEN
Key points
• The Cmr complex is a spiral large molecular complex and is composed of six different protein subunits and an RNA.
• The Cmr complex cleaves viral RNAs at five different sites, thereby destroying the virus.
• Research results contribute to clarifying the origin and evolution of the complicated acquired immune systems of higher animals.
|
RIKEN (President, Ryoji Noyori) has clarified the structure and function of the Cmr complex, which is a large complex playing a central role in bacterial immune systems. This discovery will be a key to elucidating the origin and evolution of the immunity that higher animals have acquired during their evolution. The achievement of this research was realized by the joint research group led by Akeo Shinkai* (senior research scientist) and Yoshihiro Agari (research fellow) from the Research Infrastructure Group, RIKEN SPring-8 Center (Director, Tetsuya Ishikawa) (*currently, a senior research scientist at the Structural Biology Laboratory led by Shigeyuki Yokoyama); Saori Maki (research scientist) from the Bio-specimen Platform Group, RIKEN SPring-8 Center; Koji Yonekura (associated chief scientist) from the Biostructural Mechanism Laboratory, RIKEN SPring-8 Center; and Raymond H. J. Staals (research scientist) and John van der Oost (professor) from Wageningen University in the Netherlands. All living organisms have immune systems that protect their bodies from external toxic substances and pathogens. There are various types of immune system in different species that are considered to have evolved along with the living organisms. Among the immune systems, acquired immunity includes advanced mechanisms, such as for memorizing information on antigens including viruses; however, both the origin and evolution process of acquired immunity have remained unclear. Bacteria are living organisms that first appeared in the early stages of the world’s natural history. Many bacteria have a host defense system called the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas system*1, which is similar to an acquired immunity system. The joint research group have been trying to clarify the entire mechanism of this system using Thermus thermophilus HB8*2 as a bacterial model to elucidate the origin and evolution of acquired immunity. This time, the group isolated the Cmr complex, which is composed of six different Cas proteins*3 and an RNA fragment and plays a central role in the CRISPR-Cas system. Then, they examined in detail the electron microscopy structure and activities of the complex. The results revealed that the Cmr complex forms a unique spiral structure and its RNA fragment binds to part of a viral RNA to trap the virus. It then cleaves the RNA to destroy the virus and prevent infection. Since the early stages of life on earth, living organisms have continued to fight against viruses. This research provides key findings that may clarify the origin and evolution of the antivirus immune and defense systems of living organisms that have evolved over a long period. This research was supported by Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Science (Grant No. 25440013). The results were published online in the American scientific journal Molecular Cell on 10 October 2013. Publication: |
<<Figures>>
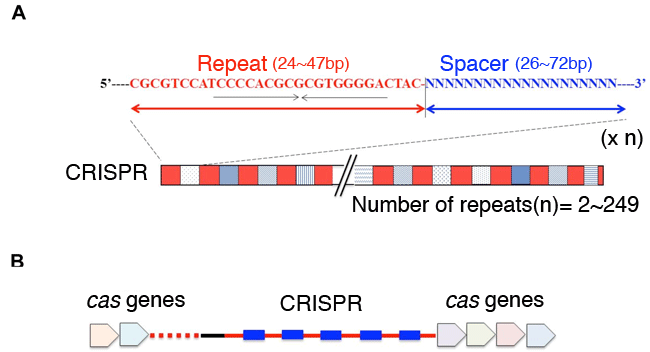
(A) CRISPR loci consist of ~25-50 bp palindromic repeat sequences (in red), separated by 25-50 bp spacer sequences (in blue). The spacers have no common features in their sequences. The maximum number of repeats is as high as 249.
(B) Cas genes are located close to the CRISPR loci.
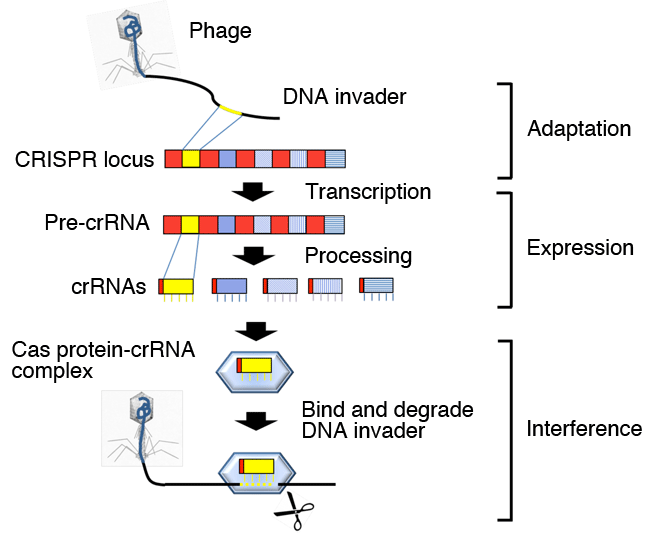
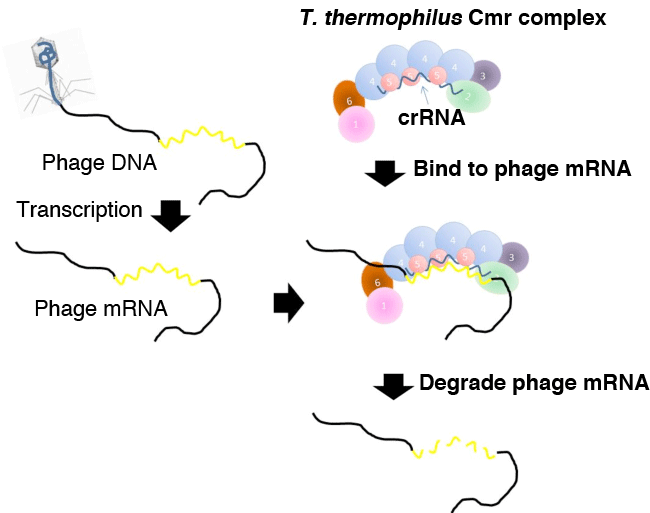
The CRISPR-Cas system has three main phases. Parts of DNAs invaded into cells such as those of viruses (phages) are cleaved, and incorporated into the CRISPR locus as a new spacer (adaptation phase). The CRISPR locus is transcribed to generate pre-crRNA, which is cleaved to generate crRNAs (expression phase). The crRNA*4 forms a complex with Cas proteins, and the complex containing the crRNA derived from the DNA invader binds to the complementary sequence of the DNA invader to degrade it (interference phase).
The number of CRISPR loci, their sequences, and the type of cas gene vary among species. Some types of CRISPR-Cas system degrade the RNAs transcribed from DNA invaders.
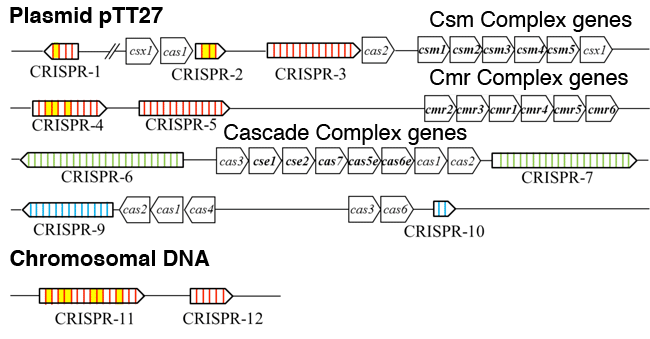
T. thermophilus HB8 has nine CRISPR loci on the plasmid pTT27 and two CRISPR loci on the chromosome (here, CRISPR-8 is excluded). These loci are classified into three types (indicated in red, green, and blue) depending on the nucleotide sequence of the repeat. Cas genes belonging to subtypes III-A (Csm)*6, III-B (Cmr), and I-E (Cascade)*5 are in bold letters.
From the analysis results, many crRNAs (in yellow) from particular spacers in CRISPR-1, CRISPR-2, CRISPR-4, and CRISPR-11 were found in the crRNA sequences of the Cmr complex. This indicates that the crRNAs selectively, rather than randomly, bind to the Cmr complex.
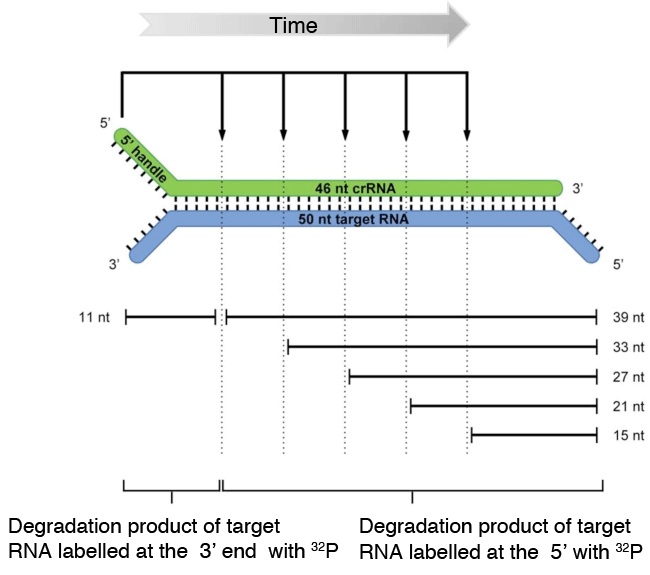
The Cmr complex binds to a 50 nt target RNA that contains complementary sequence of the 46 nt crRNA contained in the Cmr complex. Then, the target RNA is cleaved from the 3’ end at five different sites at 6 nt intervals.
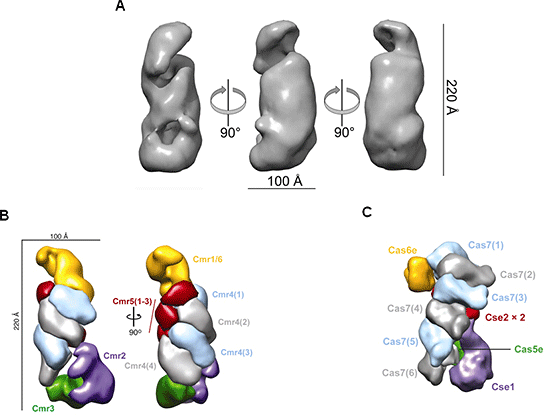
(A) Structures of T. thermophilus Cmr complexes obtained by electron microscopy.
(B) Cas protein subunit composition of the Cmr complex reconstruction based on the results of mass spectrometry*7 and electron microscopy. The stoichiometry is Cmr1:Cmr2:Cmr3:Cmr4:Cmr5:Cmr6 = 1:1:1:4:3:1. Four Cmr4 and three Cmr5 subunits form a spiral structure.
© Cas protein subunit composition of the E. coli Type I-E (Cascade) complex reconstruction based on the structure obtained by electron microscopy. Six Cas7 subunits form a spiral structure.
The Cmr complex binds to -viral (phage) mRNA at the sequence that is complementary to the crRNA of the Cmr complex, and cleaves it at five different sites. Four Cmr4 subunits located at the center of the Cmr complex and another type of Cmr subunit are possibly have active sites for cleavage.
<<Glossary>>
*1 CRISPR-Cas system
Bacterial immune and defense systems against invading nucleic acids, such as viruses (phages) and plasmids. The CRISPR loci consist of ~25-50 bp palindromic DNA repeats, separated by ~25-50 bp sequences (spacers). Cas proteins are encoded in gene clusters close to these CRISPR loci.
*2 Thermus thermophilus HB8
A bacterium found in Mine hot spring in Izu peninsula, Shizuoka Prefecture, Japan. This strain can grow at the extremely high temperature of 85 °C. Thermophilic bacteria including T. thermophilus HB8 have been thought to be close to the common ancestor of all living organisms and have the basic characteristics of primitive life. T. thermophilus HB8 has 11 CRISPR loci and ~30 different cas genes.
*3 Cas protein
Cas proteins are encoded in the genes close to the CRISPR loci. So far, 45 families have been found. Cas proteins in some families form a large molecular complex comprising several types of subunit. Cas proteins are involved in the immune and defense systems formed by the CRISPR-Cas complex.
*4 CRISPR RNA (crRNA)
crRNA binds to Cas proteins to form a complex. This complex binds to the complementary sequence of DNA or RNA invaders and degrades them.
*5 Type I-E (Cascade) complex
The Cascade complex is composed of Cse1, Cse2, Cas7, Cas5e, and Cas6e proteins and crRNA and forms a 405 kDa complex with a stoichiometry of 1:2:6:1:1:1. The Cascade complex cleaves pre-crRNA to generate crRNA and incorporates it. In addition, the complex binds to the Cas3 protein to degrade a target DNA. The detailed functional mechanism and electron microscopy structure of the Escherichia coli Cascade complex have been clarified.
*6 Type III-A (Csm) complex
The Csm complex is composed of five different protein subunits (Csm1, Csm2, Csm3, Csm4, and Csm5) and crRNA, and is associated with the cleavage of DNA.
*7 Mass spectrometry
When a sample is ionized by a high voltage in an apparatus under vacuum, the ions move around in the apparatus owing to the electrostatic force. The moving ions can be separated according to their mass-to-charge ratio using electric and magnetic fields. Different types of ion are then detected independently to obtain mass spectra in which the mass-to-charge ratio is on the x-axis and the detected intensity is on the y-axis. The molecular weight of the sample can be determined from the spectra.
|
For more information, please contact: |
- Previous Article
- Development of a methodology to control polymer orientation - improving solar cell efficiency (Press Release)
- Current article
- Exploration of Origin of Acquired Immunity (Press Release)