Successful Use of Artificial Capsule for Capturing a Protein Alive (Press Release)
- Release Date
- 03 Oct, 2012
- BL38B1 (Structural Biology III)
- BL41XU (Structural Biology I)
Japan Science and Technology Agency (JST)
The University of Tokyo, School of Engineering
Institute for Molecular Science (National Institutes of Natural Science)
RIKEN
Japan Synchrotron Radiation Research Institute
Main points
• The world’s first artificial precision capsule capable of containing huge molecules, e.g. proteins.
• “Self-organization” utilized for the preparation: simple mixing of metallic ions and organic compounds triggers a spontaneous organization process.
• Wide use expected in various areas in industries and drug discovery, typically for structure analysis and functional modification of proteins.
|
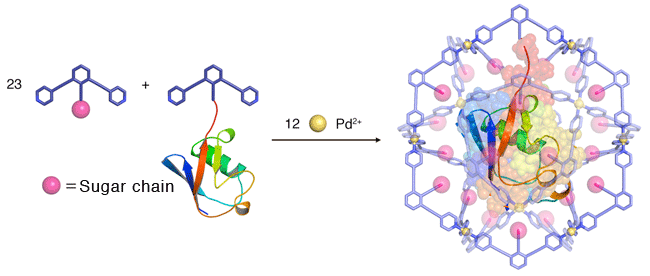
The researchers, including Prof. Makoto Fujita (Department of Applied Chemistry, School of Engineering, the University of Tokyo ) and Prof Koichi Kato (Okazaki Institute of Integrative Bioscience, JST), conducted research on the artificial formation of a nanoscale capsule as part of JST Mission-oriented Basic Research, and succeeded in trapping a whole protein molecule inside a capsule with a diameter of seven nanometers (10-9 m). There are many well-known systems in nature in which a biological molecule, e.g. a protein or DNA, is contained within a capsule structure, such as a virus shell. The capsule controls the structure and activities of the molecule within it, or stores it until a condition arises to break dormancy. In artificially prepared chemical phenomena, certain hollow molecules, or a host, are known to contain smaller organic molecules and control their structures and activities. Up to the present, however, the upsizing of the artificially synthesized molecular capsules with a precisely-controlled structure has been presented with many difficulties, inhibiting them from containing a large biological molecule, such as a protein. The research team prepared a metal complex*1 (an artificial capsule) with the aid of the “self-organization” phenomenon - a spontaneous process of generating an ordered structure triggered by the mixing of metallic ions and certain types of engineered organic compounds - and succeeded in containing protein as a whole within it. The molecular system was then crystallized and served for X-ray crystal diffraction analysis: Dr. Masaki Takata (Deputy director, RIKEN SPring-8 Center) and Dr. Takashi Kumasaka (vice chief researcher, JASRI) analyzed the molecular structure based on the X-ray diffraction data obtained from the structural biology beam lines (BL41XU, BL38B1) at SPring-8 and the PF-AR NE3A beam line at the KEK Photon Factory, and revealed the characteristic configuration, i.e. a protein molecule wrapped inside an artificial capsule. The group led by Dr. Susumu Uchiyama (associate professor at Osaka University, Division of Advanced Science and Biotechnology) also analyzed the molecular weight of the system in solution state using ultracentrifuge analysis,*2 and proved that the protein is stably contained in the capsule even in a solution. The technique used in this study is applicable in precision synthesis of artificial capsules capable of wrapping around a bulky biological molecule as a whole, creating an expectation for contributing industries, typically in drug discovery. The research result was published in the online version of Nature Communications (a UK science journal) on October 2, 2012 (UKT). Publication: |
<<Figures>>
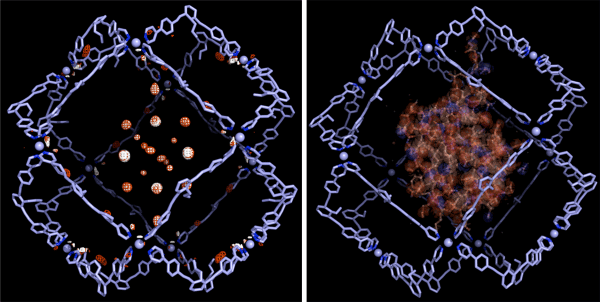
(Left) The electron density mapping of the protein as revealed by the single-crystal structure analysis*4 method in combination with MEM*3.
(Right) The steric structure of the protein obtained by simulation.
<<Glossary>>
*1 Metal complex
Molecules consisting of metal ions and ligands.
*2 Ultracentrifuge analysis
A measurement method to track molecular behaviors, including molecular weight distribution, and configuration, in a solution state under an extreme centrifugal force as large as 2x105 g. The results of the ultracentrifuge analysis in this study indicated a unique molecular weight distribution with a positive shift by the amount of the molecular weight of the connected ubiquitin.
*3 MEM (Maximum Entropy Method)
The maximum entropy method (MEM) is a data processing technique that tries to obtain information as accurate as possible from the limited amount of experimental data. MEM as applied to this study proved effective for quantitative evaluation of the weak electron density within the void and for visualizing it.
*4 Single-crystal structure analysis
A method to determine molecular structure which involves X-ray irradiation of a crystallized sample and analysis of diffracted beams, or reflective spots. The method as applied to this study proved effective in determining the steric molecular structure of the capsule molecules that contained proteins.
*5 Sugar chain
The sugar chain has many intra-molecular hydroxyl groups that render it highly hydrophilic, thus it is expected to show affinity to protein surfaces.
|
For more information, please contact:
Prof. Koichi Kato (Department of Life and Coordination-Complex Molecular Science) |
- Previous Article
- Successful Observation of “Solid-like” Molecular Behaviors in Supercooled Liquid (Press Release)
- Current article
- Successful Use of Artificial Capsule for Capturing a Protein Alive (Press Release)