Topic 3: Real-Time Analysis of Clock Protein using Small-Angle X-ray Scattering (SAXS)
Clarification of Mechanisms How the “Biological Clock” of Cyanobacteria Ticks
Many organisms on Earth have precise biological clocks; humans also possess an intrinsic chronometer, which acts like a metronome for our biological functions. Organisms conduct essential biological functions, such as metabolism and photosynthesis, according to a circadian rhythm with a 24-hour cycle. A group of proteins called “clock proteins” control these biological clocks. Numerous species of organisms are used to research these clock proteins. Highly brilliant X-rays at SPring-8 can visualize real-time changes in clock proteins of cyanobacteria (the simplest form of organisms with biological clocks), which had never been expected to be possible. This achievement will open up a new dimension for the basic research on biological clocks, including the relationship between biological clocks and diseases.
The Principle of “Clocks” is the Assembly and Disassembly of Three Types of Proteins
Clock functions of cyanobacteria are governed by proteins called “Kai.” There are three types of Kai proteins: KaiA, KaiB, and KaiC. KaiC plays a role of the “clock pendulum,” and its state varies according to its interactions with the other two Kai proteins.
Adenosine triphosphate (ATP) plays an important role in these interactions. ATP is an essential component that organisms use to store and utilize energy; it is often referred to as the “energy currency” of the cell. Energy is stored when adenosine diphosphate (ADP) is converted into ATP, and released when ATP is converted into ADP. In a process called phosphorylation, ATP-derived phosphates are transferred to specific proteins, modifying their structures and functions.
In cyanobacteria, KaiA induces phosphorylation of KaiC, and KaiB suppresses the functions of KaiA, which induces dephosphorylation of KaiC. KaiC is “oscillating” through phosphorylation and dephosphorylation by using ATP as an energy source. It is an amazing biological phenomenon that only three types of proteins govern the stable ticking of this cellular clock.
To elucidate this astonishing mechanism, the structures of the Kai proteins need to be revealed. The 3D structure of each Kai protein has been individually determined, but the structure of their composite form has yet to be explored. The difficulty lies in the fact that the complex is constantly changing its size and shape, instead of maintaining a stable binding state. Since such assembly and disassembly phenomena cannot be observed by static measurements, the existing techniques of structural biology is of only limited use for this purpose.
Sophisticated Collaborative Work between KaiA and KaiB
In October 2005, Dr. Shuji Akiyama1) (a researcher in the Precursory Research for Embryonic Science and Technology funded by the Japan Science and Technology Agency) and Dr. Yuichiro Maeda2) (Chief Scientist at RIKEN, Japan) led a research group to address the challenge of studying the Kai complex using the RIKEN Structural Biology I beamline (BL45XU).
First of all, KaiA, KaiB, and KaiC were incubated with ATP to initiate the phosphorylation cycle. A constant quantity of samples was extracted from this incubation solution every three hours. The fraction of KaiC that was phosphorylated by ATP was quantitated using a portion of this extracted sample solution.
The remainder of each extracted sample solution was irradiated with X-rays at BL45XU for 72 hours, and small-angle X-ray scattering (SAXS) data were taken. SAXS is an experimental technique in which scattered X-rays are detected at very low angles to obtain information about the structures of samples at the level of a few nanometers (1 nm = 10-9 m). Scattered X-rays were captured with X-ray cameras, and the intensity and angular distribution of scattered X-rays were obtained from the recorded X-ray images in order to determine the composition of the structures of the Kai proteins.
These measurements revealed that the intensity of the scattered X-rays in the forward direction (scattering intensity at the origin) was robustly oscillating with a 24-hour cycle. This indicates that the Kai proteins are continually assembling and disassembling in solution. Additionally, it was revealed that the phosphorylation state of KaiC was oscillating with a 24-hour cycle (Fig. 1). “Interestingly, the phosphorylation process preceded the timing of the increase and decrease of the weighted-average molecular weight (Mw) of the sample by a quarter cycle. We conducted similar experiments on the system lacking KaiA and KaiB to investigate the cause of this phase difference,” said Dr. Akiyama.
The timing of the interactions between KaiC and KaiA significantly differs from that of between KaiC and KaiB, which proceeds in the following manner. When the three types of Kai proteins are mixed, KaiA first quickly binds KaiC and phosphorylates it. After the phosphorylation of KaiC is completed, KaiB slowly binds KaiC to dephosphorylate it, and then dissociates from KaiC. The combination of the quick process involving KaiA and the slow process involving KaiB induces the phase difference.
Furthermore, this research group elucidated the structures of the KaiA-KaiC and KaiB-KaiC complexes, which are produced during assembly and disassembly processes, by examining the molecular shape of these two complexes based on the information obtained from the SAXS measurement (Fig. 2).
The size of the KaiA-KaiC complex is 15 × 15 × 11 nm. KaiC occupies 80% of the total volume and consists of a spherical head region, which has a cavity in the center, and a short tail region. The head region of KaiC is divided into two domains: C1 and C2. The remaining 20% of the complex, which corresponds to KaiA, is clustered near the tail.
The size of the KaiB-KaiC complex measures 14 × 13 × 10 nm, slightly smaller than the KaiA-KaiC complex. Similar to the KaiA-KaiC complex, the structure characteristic to KaiC can be observed in the KaiB-KaiC complex. KaiB, which occupies 13% of the entire volume, is acting on the C2 domain.
These molecular structures discovered through SAXS are consistent with the structures of individual Kai proteins that were determined by other research groups; therefore, it can be concluded that the determination of the complex structures proceeded smoothly. Finally, Dr. Akiyama and colleagues have used visualization methods to elucidate, for the first time, the real-time assembly/disassembly dynamics of the Kai proteins. The results of their studies on the crystal structures of the biological clocks of cyanobacteria was published in Molecular Cell (March 2008).
1) Currently Associate Professor at Nagoya University, Japan.
2) Currently Professor at Nagoya University, Japan.
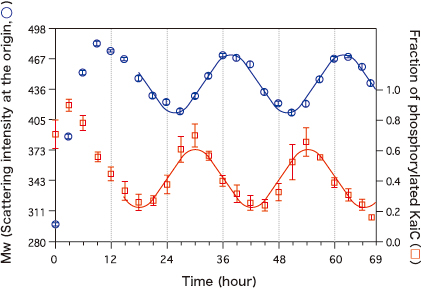
The left axis represents the weighted-average molecular weight (Mw) of samples, estimated from the intensity of the scatted X-rays (scattering intensity at the origin); the right axis represents the fraction of phosphorylated KaiC (phosphorylated state) measured at the same time. The phosphorylated state precedes Mw by a quarter cycle.
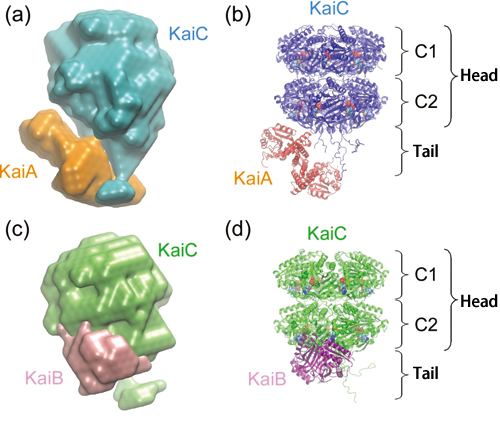
KaiA and KaiB interact with the C2 domain and tail region of KaiC.
True Identity of the “Clock Pacemaker”
“We identified gears that play the role of pacemakers to control the ticking of the biological clock. We believe this is a more important achievement than the visualization,” said Dr. Akiyama. The next question then, is “Where is the pacemaker?”
This research group paid attention to the fact that amino acids that are to be phosphorylated are localized in the C2 domain of KaiC; the interactions with KaiA and KaiB are induced in the C2 domain and tail region of KaiC as well. These facts indicate that KaiC alters the structures of the C2 domain and tail region, according to its own phosphorylation state, to pull or repel KaiA and KaiB and then to control the timing of the reactions (Fig. 3). KaiC, which seems to be driven by KaiA and KaiB, is actually the pacemaker.
How can KaiC keep such a precise timing? To answer this question, it is necessary to elucidate the mechanisms of the structure changes of KaiC in solutions. This research group, now armed with SAXS, will investigate the mechanisms by which KaiC exerts its functions as a pacemaker. The mechanisms of the biological clocks of humans will also be elucidated eventually. When that is achieved, the development of breakthrough therapies for sleep disorders caused by biological rhythm dysfunction will no longer be a dream.
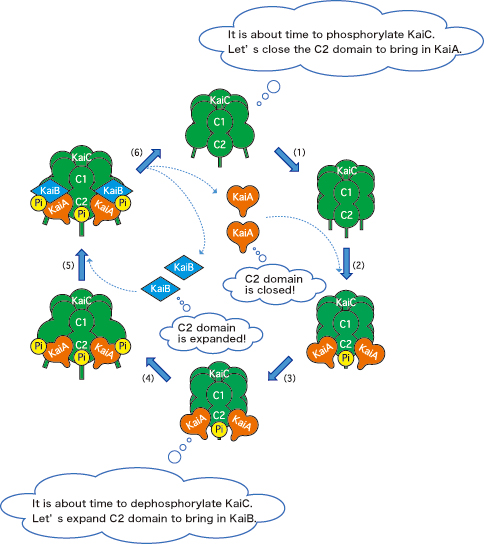
KaiA, orange heart; KaiB, blue diamond; KaiC, green barrel; phosphate, “Pi.” (1) KaiC alters the C2 domain structure to bring in KaiA. (2) KaiA binds KaiC. (3) KaiC is phosphorylated. (4) KaiC further alters the C2 domain structure to bring in KaiB. (5) KaiB binds the KaiA-KaiC complex. (6) KaiC is dephosphorylated; KaiA and KaiB dissociate from KaiC.
Reference
1. S. Akiyama, A. Nohara, K. Ito and Y. Maéda; Molecular Cell, 29, 703 (2008)