Clarification of Structure of Actin Filament by X-Ray Fiber Diffraction - Step towards clarifying mechanism behind physiological functions performed by vital fundamental protein, "actin" (Press Release)
- Release Date
- 22 Jan, 2009
- BL41XU (Structural Biology I)
- BL40B2 (Structural Biology II)
- BL45XU (RIKEN Structural Biology I)
RIKEN
Japan Science and Technology Agency
Nagoya University
Key research achievements
- Verification of structural model and analysis results for actin mutant, establishing a new detailed structural model
- Necessity of the flattening of actin molecules from monomers to polymers to form a filament
- Obtaining a key to understanding all types of vital phenomena in a cell
Researchers at RIKEN (Ryoji Noyori, President), the Japan Science and Technology Agency (JST; Koichi Kitazawa, President), and Nagoya University (Shin-ichi Hirano, President) clarified the detailed structure of actin filaments. Actin, a fundamental protein that supports life-sustaining functions, is polymerized to form a chain of actin monomers, which is called an actin filament. It was found that two large twisted domains of an actin monomer rotate with respect to each other and form a platelike structure during polymerization. This finding will hopefully unveil the mechanism underlying cell motility associated with actin. This was achieved in a joint research project by Toshiro Oda, leader of the X-Ray Structural Analysis Research Team, RIKEN SPring-8 Center, and Tomoki Aihara, a researcher of Dr. Oda's team; Yuichiro Maeda, a professor, and Tetsuhiro Narita, an associate professor of the Structural Biology Research Center, Graduate School of Science, Nagoya University; and Mitsusada Iwasa, a researcher of the "Maeda Actin-Filament Dynamics Project" supported by the Exploratory Research for Advanced Technology (ERATO) program of the JST.
Actin is a protein abundant in eukaryotic cells and performs fundamental physiological functions, such as cell motility, intracellular motility, organelle immobilization, and cell division. Two states of actin are known – an isolated, globular monomer (G-actin) and a filamentous polymer (F-actin). Generally, cell motility is caused by the transformation from G-actin to F-actin. After the discovery of actin in 1942, Fumio Ohsawa, professor emeritus at Nagoya University and Osaka University, and his colleagues studied actin, focusing on its polymerization from the thermodynamic aspect in the 1950s and 1960s. K.C. Holmes, the director of Max Planck Gesellschaft in Germany, and his coworkers elucidated the crystal structure of the actin monomer in 1990. A model comprising a simple stack of crystal structures of this monomer is still used as the conventional structural model of F-actin.
In this research, a structural model of F-actin was constructed by X-ray fiber diffraction using the synchrotron radiation beamlines at SPring-8, BL41XU (Structural Biology I), BL40B2 (Structural Biology II), and BL45XU (RIKEN Structural Biology I). Through the verification of the model using a cryoelectron microscope and by the analysis of an actin mutant, the researchers developed a new detailed structural model, in which the structural change due to the polymerization of actin is as simple as the rotation of two large domains relative to each other. We hope that this model will contribute to the establishment of a basis for clarifying the mechanisms underlying the physiological functions of actin, such as binding actin molecules to form a filament and binding with actin-binding proteins.
This research is part of the "Maeda Actin-Filament Dynamics Project" (Yuichiro Maeda, Research Director) supported by the ERATO program of the JST. These achievements were published in the scientific journal Nature online on 22 January 2009.
Publication:
"The nature of the globular- to fibrous-actin transition"
Toshiro Oda, Mitsusada Iwasa, Tomoki Aihara, Yuichiro Maéda & Akihiro Narita
Nature 457, 441-445 (2009), published online 22 January 2009
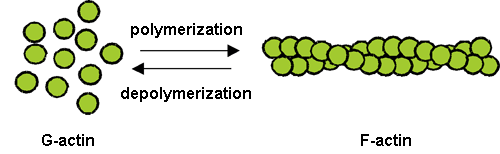 Fig. 1 Polymerization and depolymerization of actin.
Fig. 1 Polymerization and depolymerization of actin.
 Fig. 2 Superconductive electromagnet.
Fig. 2 Superconductive electromagnet.
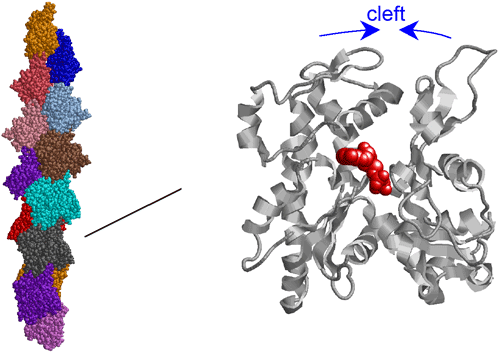 Fig. 3 Structural model of F-actin.
Fig. 3 Structural model of F-actin.
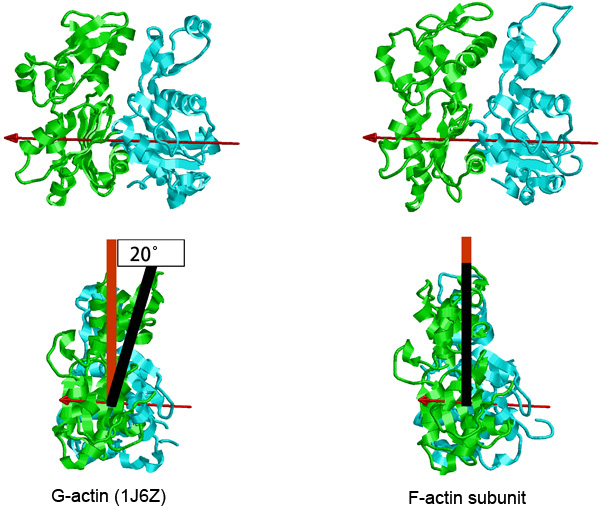 Fig. 4 G-actin and an actin molecule forming F-actin.
Fig. 4 G-actin and an actin molecule forming F-actin.
|
For more information, please contact: |
- Previous Article
- Successful Structural Determination of Vault in Higher Organisms Using SPring-8 Beamline (Press Release)
- Current article
- Clarification of Structure of Actin Filament by X-Ray Fiber Diffraction - Step towards clarifying mechanism behind physiological functions performed by vital fundamental protein, "actin" (Press Release)
 .
.